Deepta Rajan
Self-training of Machine Learning Models for Liver Histopathology: Generalization under Clinical Shifts
Nov 14, 2022

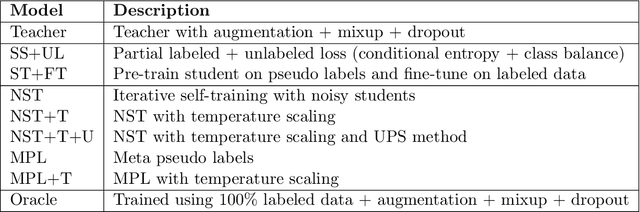

Abstract:Histopathology images are gigapixel-sized and include features and information at different resolutions. Collecting annotations in histopathology requires highly specialized pathologists, making it expensive and time-consuming. Self-training can alleviate annotation constraints by learning from both labeled and unlabeled data, reducing the amount of annotations required from pathologists. We study the design of teacher-student self-training systems for Non-alcoholic Steatohepatitis (NASH) using clinical histopathology datasets with limited annotations. We evaluate the models on in-distribution and out-of-distribution test data under clinical data shifts. We demonstrate that through self-training, the best student model statistically outperforms the teacher with a $3\%$ absolute difference on the macro F1 score. The best student model also approaches the performance of a fully supervised model trained with twice as many annotations.
Revisiting Inlier and Outlier Specification for Improved Out-of-Distribution Detection
Jul 12, 2022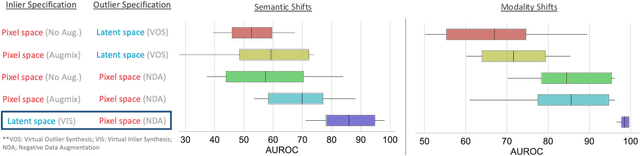
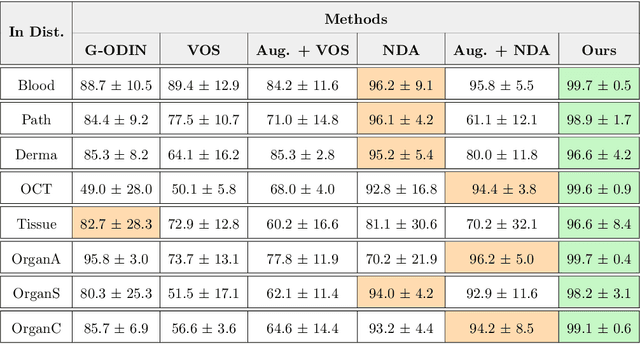
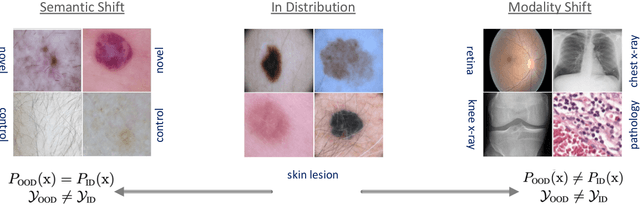
Abstract:Accurately detecting out-of-distribution (OOD) data with varying levels of semantic and covariate shifts with respect to the in-distribution (ID) data is critical for deployment of safe and reliable models. This is particularly the case when dealing with highly consequential applications (e.g. medical imaging, self-driving cars, etc). The goal is to design a detector that can accept meaningful variations of the ID data, while also rejecting examples from OOD regimes. In practice, this dual objective can be realized by enforcing consistency using an appropriate scoring function (e.g., energy) and calibrating the detector to reject a curated set of OOD data (referred to as outlier exposure or shortly OE). While OE methods are widely adopted, assembling representative OOD datasets is both costly and challenging due to the unpredictability of real-world scenarios, hence the recent trend of designing OE-free detectors. In this paper, we make a surprising finding that controlled generalization to ID variations and exposure to diverse (synthetic) outlier examples are essential to simultaneously improving semantic and modality shift detection. In contrast to existing methods, our approach samples inliers in the latent space, and constructs outlier examples via negative data augmentation. Through a rigorous empirical study on medical imaging benchmarks (MedMNIST, ISIC2019 and NCT), we demonstrate significant performance gains ($15\% - 35\%$ in AUROC) over existing OE-free, OOD detection approaches under both semantic and modality shifts.
Designing Counterfactual Generators using Deep Model Inversion
Oct 05, 2021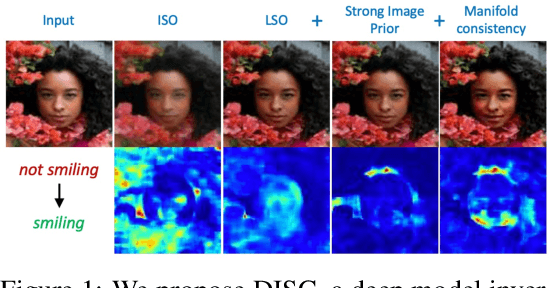
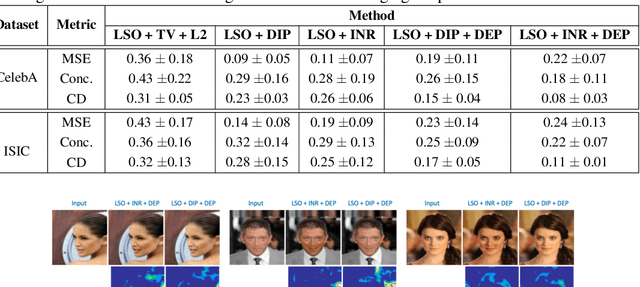
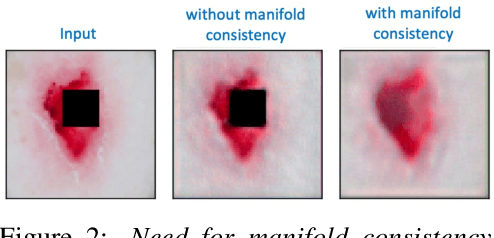

Abstract:Explanation techniques that synthesize small, interpretable changes to a given image while producing desired changes in the model prediction have become popular for introspecting black-box models. Commonly referred to as counterfactuals, the synthesized explanations are required to contain discernible changes (for easy interpretability) while also being realistic (consistency to the data manifold). In this paper, we focus on the case where we have access only to the trained deep classifier and not the actual training data. While the problem of inverting deep models to synthesize images from the training distribution has been explored, our goal is to develop a deep inversion approach to generate counterfactual explanations for a given query image. Despite their effectiveness in conditional image synthesis, we show that existing deep inversion methods are insufficient for producing meaningful counterfactuals. We propose DISC (Deep Inversion for Synthesizing Counterfactuals) that improves upon deep inversion by utilizing (a) stronger image priors, (b) incorporating a novel manifold consistency objective and (c) adopting a progressive optimization strategy. We find that, in addition to producing visually meaningful explanations, the counterfactuals from DISC are effective at learning classifier decision boundaries and are robust to unknown test-time corruptions.
Loss Estimators Improve Model Generalization
Mar 05, 2021

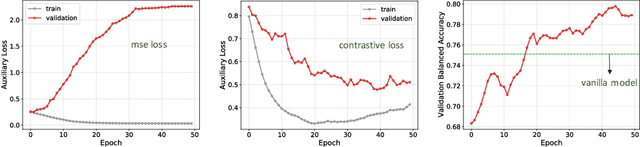

Abstract:With increased interest in adopting AI methods for clinical diagnosis, a vital step towards safe deployment of such tools is to ensure that the models not only produce accurate predictions but also do not generalize to data regimes where the training data provide no meaningful evidence. Existing approaches for ensuring the distribution of model predictions to be similar to that of the true distribution rely on explicit uncertainty estimators that are inherently hard to calibrate. In this paper, we propose to train a loss estimator alongside the predictive model, using a contrastive training objective, to directly estimate the prediction uncertainties. Interestingly, we find that, in addition to producing well-calibrated uncertainties, this approach improves the generalization behavior of the predictor. Using a dermatology use-case, we show the impact of loss estimators on model generalization, in terms of both its fidelity on in-distribution data and its ability to detect out of distribution samples or new classes unseen during training.
Automatic Diagnosis of Pulmonary Embolism Using an Attention-guided Framework: A Large-scale Study
May 29, 2020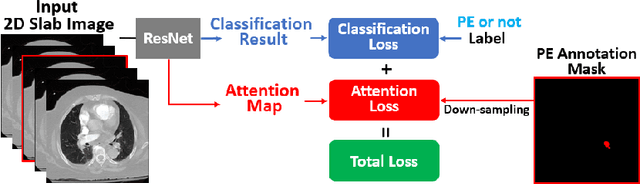
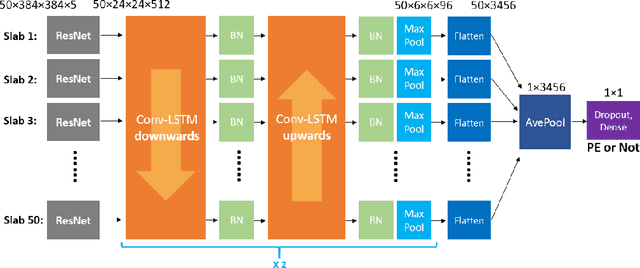
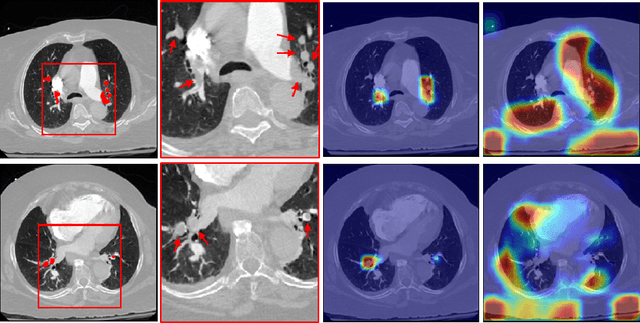
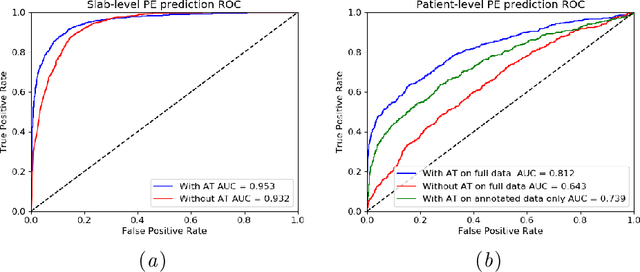
Abstract:Pulmonary Embolism (PE) is a life-threatening disorder associated with high mortality and morbidity. Prompt diagnosis and immediate initiation of therapeutic action is important. We explored a deep learning model to detect PE on volumetric contrast-enhanced chest CT scans using a 2-stage training strategy. First, a residual convolutional neural network (ResNet) was trained using annotated 2D images. In addition to the classification loss, an attention loss was added during training to help the network focus attention on PE. Next, a recurrent network was used to scan sequentially through the features provided by the pre-trained ResNet to detect PE. This combination allows the network to be trained using both a limited and sparse set of pixel-level annotated images and a large number of easily obtainable patient-level image-label pairs. We used 1,670 sparsely annotated studies and more than 10,000 labeled studies in our training. On a test set with 2,160 patient studies, the proposed method achieved an area under the ROC curve (AUC) of 0.812. The proposed framework is also able to provide localized attention maps that indicate possible PE lesions, which could potentially help radiologists accelerate the diagnostic process.
Self-Training with Improved Regularization for Few-Shot Chest X-Ray Classification
May 03, 2020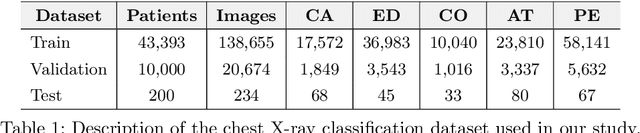
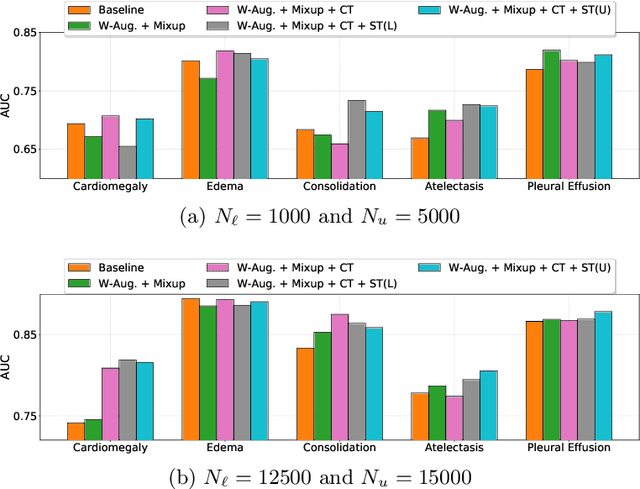
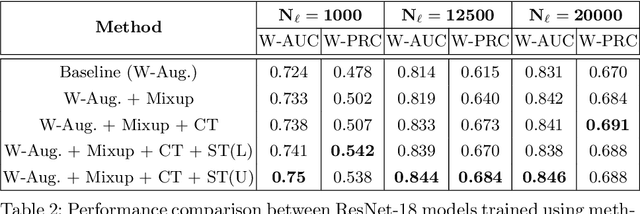
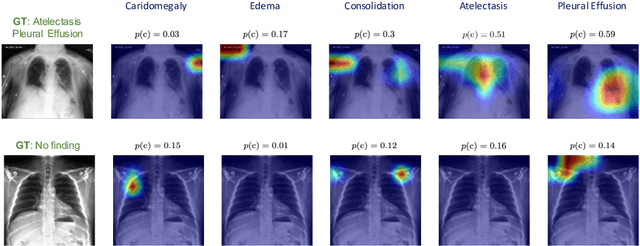
Abstract:Automated diagnostic assistants in healthcare necessitate accurate AI models that can be trained with limited labeled data, can cope with severe class imbalances and can support simultaneous prediction of multiple disease conditions. To this end, we present a novel few-shot learning approach that utilizes a number of key components to enable robust modeling in such challenging scenarios. Using an important use-case in chest X-ray classification, we provide several key insights on the effective use of data augmentation, self-training via distillation and confidence tempering for few-shot learning in medical imaging. Our results show that using only ~10% of the labeled data, we can build predictive models that match the performance of classifiers trained in a large-scale data setting.
Calibrating Healthcare AI: Towards Reliable and Interpretable Deep Predictive Models
Apr 27, 2020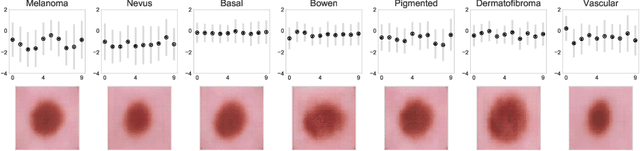
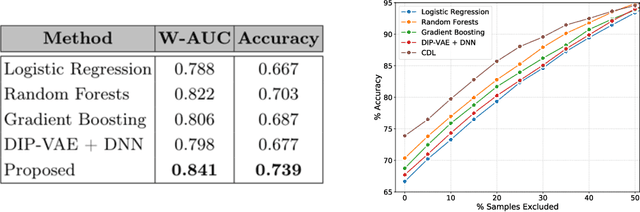
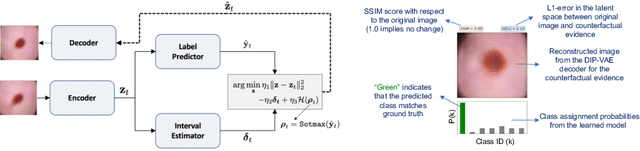
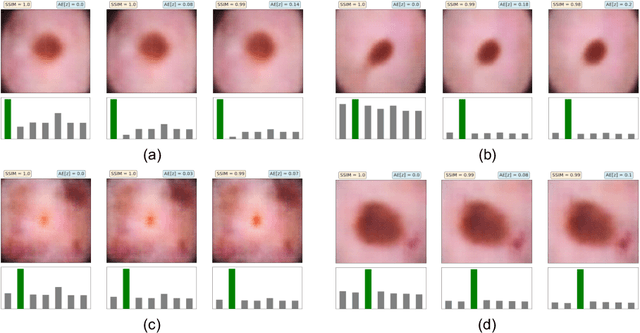
Abstract:The wide-spread adoption of representation learning technologies in clinical decision making strongly emphasizes the need for characterizing model reliability and enabling rigorous introspection of model behavior. While the former need is often addressed by incorporating uncertainty quantification strategies, the latter challenge is addressed using a broad class of interpretability techniques. In this paper, we argue that these two objectives are not necessarily disparate and propose to utilize prediction calibration to meet both objectives. More specifically, our approach is comprised of a calibration-driven learning method, which is also used to design an interpretability technique based on counterfactual reasoning. Furthermore, we introduce \textit{reliability plots}, a holistic evaluation mechanism for model reliability. Using a lesion classification problem with dermoscopy images, we demonstrate the effectiveness of our approach and infer interesting insights about the model behavior.
Learn-By-Calibrating: Using Calibration as a Training Objective
Oct 30, 2019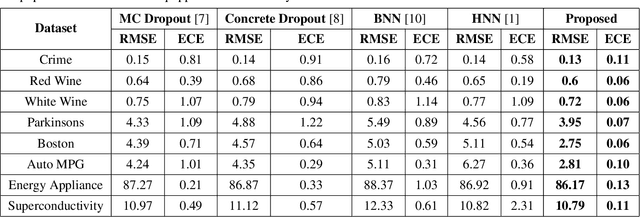
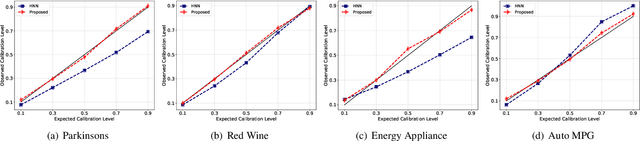
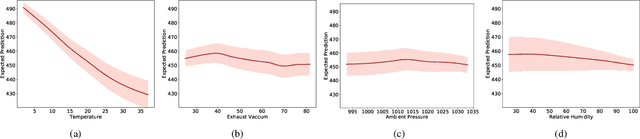
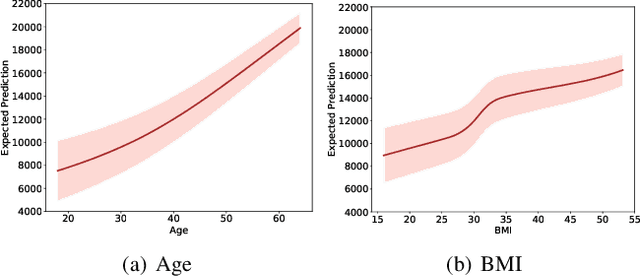
Abstract:Calibration error is commonly adopted for evaluating the quality of uncertainty estimators in deep neural networks. In this paper, we argue that such a metric is highly beneficial for training predictive models, even when we do not explicitly measure the uncertainties. This is conceptually similar to heteroscedastic neural networks that produce variance estimates for each prediction, with the key difference that we do not place a Gaussian prior on the predictions. We propose a novel algorithm that performs simultaneous interval estimation for different calibration levels and effectively leverages the intervals to refine the mean estimates. Our results show that, our approach is consistently superior to existing regularization strategies in deep regression models. Finally, we propose to augment partial dependence plots, a model-agnostic interpretability tool, with expected prediction intervals to reveal interesting dependencies between data and the target.
Pi-PE: A Pipeline for Pulmonary Embolism Detection using Sparsely Annotated 3D CT Images
Oct 21, 2019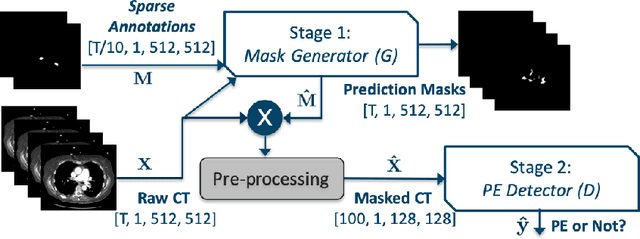
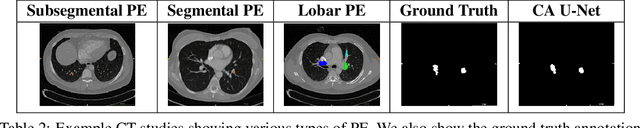
Abstract:Pulmonary embolisms (PE) are known to be one of the leading causes for cardiac-related mortality. Due to inherent variabilities in how PE manifests and the cumbersome nature of manual diagnosis, there is growing interest in leveraging AI tools for detecting PE. In this paper, we build a two-stage detection pipeline that is accurate, computationally efficient, robust to variations in PE types and kernels used for CT reconstruction, and most importantly, does not require dense annotations. Given the challenges in acquiring expert annotations in large-scale datasets, our approach produces state-of-the-art results with very sparse emboli contours (at 10mm slice spacing), while using models with significantly lower number of parameters. We achieve AUC scores of 0.94 on the validation set and 0.85 on the test set of highly severe PEs. Using a large, real-world dataset characterized by complex PE types and patients from multiple hospitals, we present an elaborate empirical study and provide guidelines for designing highly generalizable pipelines.
Leveraging Medical Visual Question Answering with Supporting Facts
May 28, 2019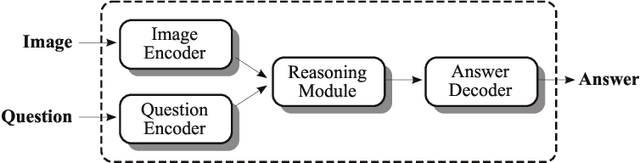
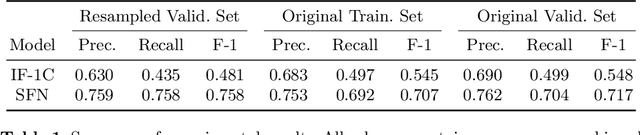
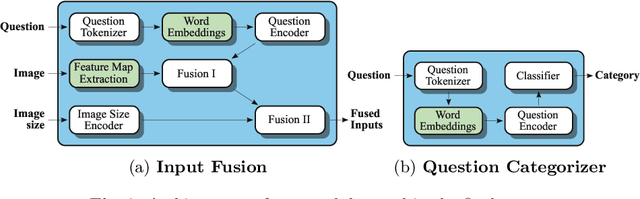
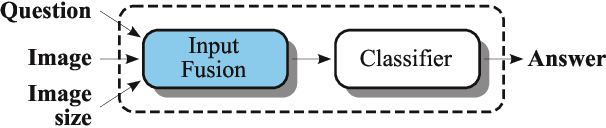
Abstract:In this working notes paper, we describe IBM Research AI (Almaden) team's participation in the ImageCLEF 2019 VQA-Med competition. The challenge consists of four question-answering tasks based on radiology images. The diversity of imaging modalities, organs and disease types combined with a small imbalanced training set made this a highly complex problem. To overcome these difficulties, we implemented a modular pipeline architecture that utilized transfer learning and multi-task learning. Our findings led to the development of a novel model called Supporting Facts Network (SFN). The main idea behind SFN is to cross-utilize information from upstream tasks to improve the accuracy on harder downstream ones. This approach significantly improved the scores achieved in the validation set (18 point improvement in F-1 score). Finally, we submitted four runs to the competition and were ranked seventh.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge