Ehsan Dehghan
Automatic Diagnosis of Pulmonary Embolism Using an Attention-guided Framework: A Large-scale Study
May 29, 2020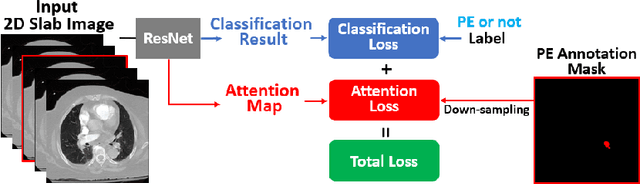
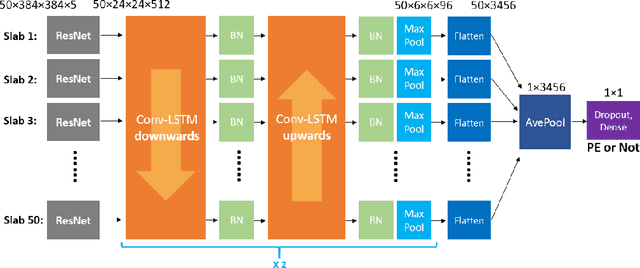
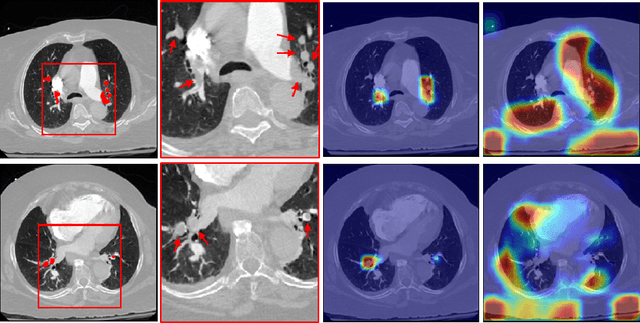
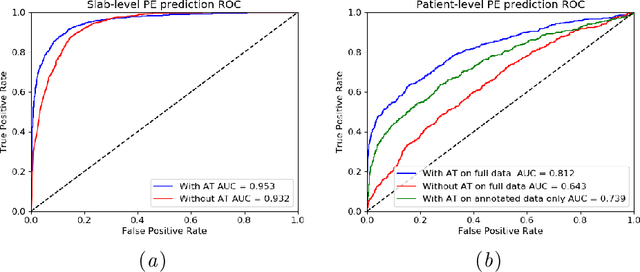
Abstract:Pulmonary Embolism (PE) is a life-threatening disorder associated with high mortality and morbidity. Prompt diagnosis and immediate initiation of therapeutic action is important. We explored a deep learning model to detect PE on volumetric contrast-enhanced chest CT scans using a 2-stage training strategy. First, a residual convolutional neural network (ResNet) was trained using annotated 2D images. In addition to the classification loss, an attention loss was added during training to help the network focus attention on PE. Next, a recurrent network was used to scan sequentially through the features provided by the pre-trained ResNet to detect PE. This combination allows the network to be trained using both a limited and sparse set of pixel-level annotated images and a large number of easily obtainable patient-level image-label pairs. We used 1,670 sparsely annotated studies and more than 10,000 labeled studies in our training. On a test set with 2,160 patient studies, the proposed method achieved an area under the ROC curve (AUC) of 0.812. The proposed framework is also able to provide localized attention maps that indicate possible PE lesions, which could potentially help radiologists accelerate the diagnostic process.
Pi-PE: A Pipeline for Pulmonary Embolism Detection using Sparsely Annotated 3D CT Images
Oct 21, 2019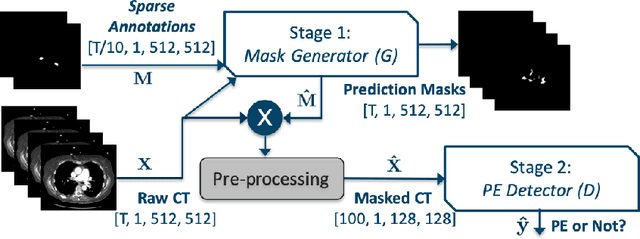
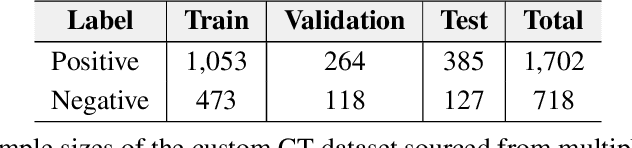
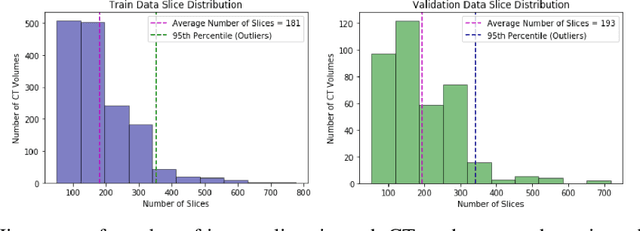
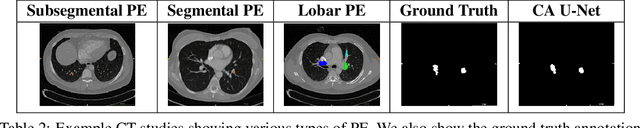
Abstract:Pulmonary embolisms (PE) are known to be one of the leading causes for cardiac-related mortality. Due to inherent variabilities in how PE manifests and the cumbersome nature of manual diagnosis, there is growing interest in leveraging AI tools for detecting PE. In this paper, we build a two-stage detection pipeline that is accurate, computationally efficient, robust to variations in PE types and kernels used for CT reconstruction, and most importantly, does not require dense annotations. Given the challenges in acquiring expert annotations in large-scale datasets, our approach produces state-of-the-art results with very sparse emboli contours (at 10mm slice spacing), while using models with significantly lower number of parameters. We achieve AUC scores of 0.94 on the validation set and 0.85 on the test set of highly severe PEs. Using a large, real-world dataset characterized by complex PE types and patients from multiple hospitals, we present an elaborate empirical study and provide guidelines for designing highly generalizable pipelines.
Disease Detection in Weakly Annotated Volumetric Medical Images using a Convolutional LSTM Network
Dec 03, 2018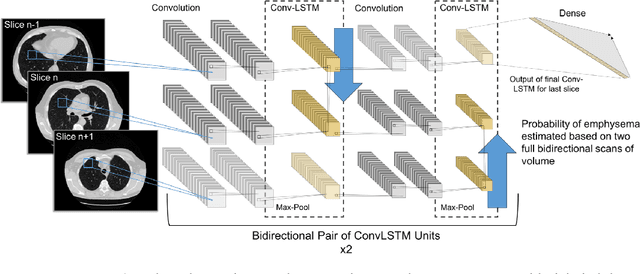
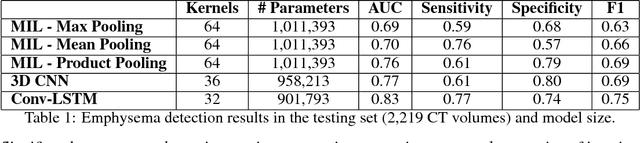
Abstract:We explore a solution for learning disease signatures from weakly, yet easily obtainable, annotated volumetric medical imaging data by analyzing 3D volumes as a sequence of 2D images. We demonstrate the performance of our solution in the detection of emphysema in lung cancer screening low-dose CT images. Our approach utilizes convolutional long short-term memory (LSTM) to "scan" sequentially through an imaging volume for the presence of disease in a portion of scanned region. This framework allowed effective learning given only volumetric images and binary disease labels, thus enabling training from a large dataset of 6,631 un-annotated image volumes from 4,486 patients. When evaluated in a testing set of 2,163 volumes from 2,163 patients, our model distinguished emphysema with area under the receiver operating characteristic curve (AUC) of .83. This approach was found to outperform 2D convolutional neural networks (CNN) implemented with various multiple-instance learning schemes (AUC=0.69-0.76) and a 3D CNN (AUC=.77).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge