Chaitanya Shivade
TN-Eval: Rubric and Evaluation Protocols for Measuring the Quality of Behavioral Therapy Notes
Mar 26, 2025Abstract:Behavioral therapy notes are important for both legal compliance and patient care. Unlike progress notes in physical health, quality standards for behavioral therapy notes remain underdeveloped. To address this gap, we collaborated with licensed therapists to design a comprehensive rubric for evaluating therapy notes across key dimensions: completeness, conciseness, and faithfulness. Further, we extend a public dataset of behavioral health conversations with therapist-written notes and LLM-generated notes, and apply our evaluation framework to measure their quality. We find that: (1) A rubric-based manual evaluation protocol offers more reliable and interpretable results than traditional Likert-scale annotations. (2) LLMs can mimic human evaluators in assessing completeness and conciseness but struggle with faithfulness. (3) Therapist-written notes often lack completeness and conciseness, while LLM-generated notes contain hallucination. Surprisingly, in a blind test, therapists prefer and judge LLM-generated notes to be superior to therapist-written notes.
CriSPO: Multi-Aspect Critique-Suggestion-guided Automatic Prompt Optimization for Text Generation
Oct 03, 2024



Abstract:Large language models (LLMs) can generate fluent summaries across domains using prompting techniques, reducing the need to train models for summarization applications. However, crafting effective prompts that guide LLMs to generate summaries with the appropriate level of detail and writing style remains a challenge. In this paper, we explore the use of salient information extracted from the source document to enhance summarization prompts. We show that adding keyphrases in prompts can improve ROUGE F1 and recall, making the generated summaries more similar to the reference and more complete. The number of keyphrases can control the precision-recall trade-off. Furthermore, our analysis reveals that incorporating phrase-level salient information is superior to word- or sentence-level. However, the impact on hallucination is not universally positive across LLMs. To conduct this analysis, we introduce Keyphrase Signal Extractor (CriSPO), a lightweight model that can be finetuned to extract salient keyphrases. By using CriSPO, we achieve consistent ROUGE improvements across datasets and open-weight and proprietary LLMs without any LLM customization. Our findings provide insights into leveraging salient information in building prompt-based summarization systems.
Entity Anchored ICD Coding
Aug 15, 2022
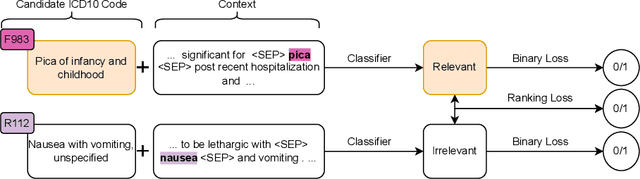
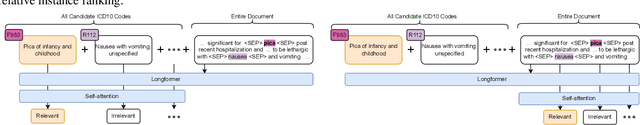
Abstract:Medical coding is a complex task, requiring assignment of a subset of over 72,000 ICD codes to a patient's notes. Modern natural language processing approaches to these tasks have been challenged by the length of the input and size of the output space. We limit our model inputs to a small window around medical entities found in our documents. From those local contexts, we build contextualized representations of both ICD codes and entities, and aggregate over these representations to form document-level predictions. In contrast to existing methods which use a representation fixed either in size or by codes seen in training, we represent ICD codes by encoding the code description with local context. We discuss metrics appropriate to deploying coding systems in practice. We show that our approach is superior to existing methods in both standard and deployable measures, including performance on rare and unseen codes.
Towards Clinical Encounter Summarization: Learning to Compose Discharge Summaries from Prior Notes
Apr 27, 2021



Abstract:The records of a clinical encounter can be extensive and complex, thus placing a premium on tools that can extract and summarize relevant information. This paper introduces the task of generating discharge summaries for a clinical encounter. Summaries in this setting need to be faithful, traceable, and scale to multiple long documents, motivating the use of extract-then-abstract summarization cascades. We introduce two new measures, faithfulness and hallucination rate for evaluation in this task, which complement existing measures for fluency and informativeness. Results across seven medical sections and five models show that a summarization architecture that supports traceability yields promising results, and that a sentence-rewriting approach performs consistently on the measure used for faithfulness (faithfulness-adjusted $F_3$) over a diverse range of generated sections.
Neural Inverse Text Normalization
Feb 12, 2021



Abstract:While there have been several contributions exploring state of the art techniques for text normalization, the problem of inverse text normalization (ITN) remains relatively unexplored. The best known approaches leverage finite state transducer (FST) based models which rely on manually curated rules and are hence not scalable. We propose an efficient and robust neural solution for ITN leveraging transformer based seq2seq models and FST-based text normalization techniques for data preparation. We show that this can be easily extended to other languages without the need for a linguistic expert to manually curate them. We then present a hybrid framework for integrating Neural ITN with an FST to overcome common recoverable errors in production environments. Our empirical evaluations show that the proposed solution minimizes incorrect perturbations (insertions, deletions and substitutions) to ASR output and maintains high quality even on out of domain data. A transformer based model infused with pretraining consistently achieves a lower WER across several datasets and is able to outperform baselines on English, Spanish, German and Italian datasets.
Receptivity of an AI Cognitive Assistant by the Radiology Community: A Report on Data Collected at RSNA
Sep 13, 2020



Abstract:Due to advances in machine learning and artificial intelligence (AI), a new role is emerging for machines as intelligent assistants to radiologists in their clinical workflows. But what systematic clinical thought processes are these machines using? Are they similar enough to those of radiologists to be trusted as assistants? A live demonstration of such a technology was conducted at the 2016 Scientific Assembly and Annual Meeting of the Radiological Society of North America (RSNA). The demonstration was presented in the form of a question-answering system that took a radiology multiple choice question and a medical image as inputs. The AI system then demonstrated a cognitive workflow, involving text analysis, image analysis, and reasoning, to process the question and generate the most probable answer. A post demonstration survey was made available to the participants who experienced the demo and tested the question answering system. Of the reported 54,037 meeting registrants, 2,927 visited the demonstration booth, 1,991 experienced the demo, and 1,025 completed a post-demonstration survey. In this paper, the methodology of the survey is shown and a summary of its results are presented. The results of the survey show a very high level of receptiveness to cognitive computing technology and artificial intelligence among radiologists.
Leveraging Medical Visual Question Answering with Supporting Facts
May 28, 2019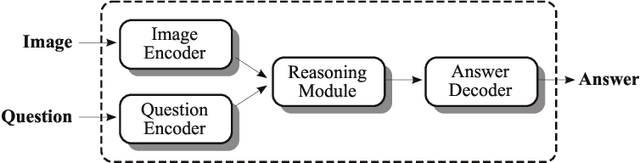
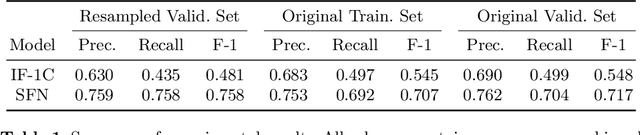
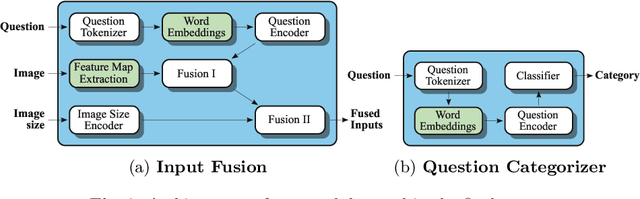
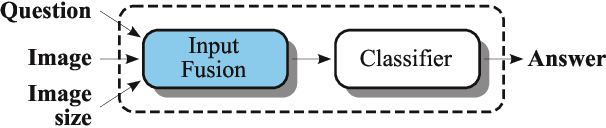
Abstract:In this working notes paper, we describe IBM Research AI (Almaden) team's participation in the ImageCLEF 2019 VQA-Med competition. The challenge consists of four question-answering tasks based on radiology images. The diversity of imaging modalities, organs and disease types combined with a small imbalanced training set made this a highly complex problem. To overcome these difficulties, we implemented a modular pipeline architecture that utilized transfer learning and multi-task learning. Our findings led to the development of a novel model called Supporting Facts Network (SFN). The main idea behind SFN is to cross-utilize information from upstream tasks to improve the accuracy on harder downstream ones. This approach significantly improved the scores achieved in the validation set (18 point improvement in F-1 score). Finally, we submitted four runs to the competition and were ranked seventh.
Towards Automatic Generation of Shareable Synthetic Clinical Notes Using Neural Language Models
May 21, 2019


Abstract:Large-scale clinical data is invaluable to driving many computational scientific advances today. However, understandable concerns regarding patient privacy hinder the open dissemination of such data and give rise to suboptimal siloed research. De-identification methods attempt to address these concerns but were shown to be susceptible to adversarial attacks. In this work, we focus on the vast amounts of unstructured natural language data stored in clinical notes and propose to automatically generate synthetic clinical notes that are more amenable to sharing using generative models trained on real de-identified records. To evaluate the merit of such notes, we measure both their privacy preservation properties as well as utility in training clinical NLP models. Experiments using neural language models yield notes whose utility is close to that of the real ones in some clinical NLP tasks, yet leave ample room for future improvements.
Lessons from Natural Language Inference in the Clinical Domain
Aug 27, 2018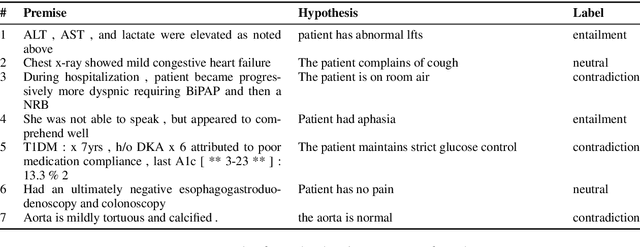

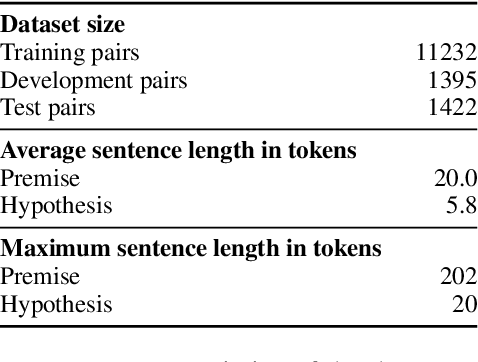
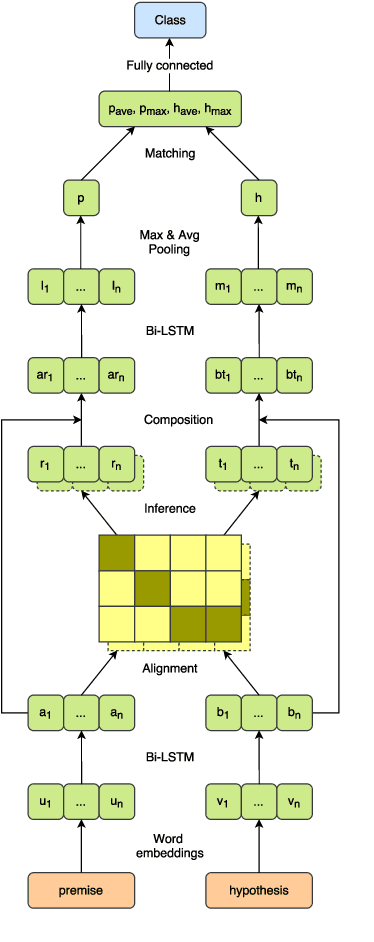
Abstract:State of the art models using deep neural networks have become very good in learning an accurate mapping from inputs to outputs. However, they still lack generalization capabilities in conditions that differ from the ones encountered during training. This is even more challenging in specialized, and knowledge intensive domains, where training data is limited. To address this gap, we introduce MedNLI - a dataset annotated by doctors, performing a natural language inference task (NLI), grounded in the medical history of patients. We present strategies to: 1) leverage transfer learning using datasets from the open domain, (e.g. SNLI) and 2) incorporate domain knowledge from external data and lexical sources (e.g. medical terminologies). Our results demonstrate performance gains using both strategies.
Addressing Limited Data for Textual Entailment Across Domains
Jun 08, 2016



Abstract:We seek to address the lack of labeled data (and high cost of annotation) for textual entailment in some domains. To that end, we first create (for experimental purposes) an entailment dataset for the clinical domain, and a highly competitive supervised entailment system, ENT, that is effective (out of the box) on two domains. We then explore self-training and active learning strategies to address the lack of labeled data. With self-training, we successfully exploit unlabeled data to improve over ENT by 15% F-score on the newswire domain, and 13% F-score on clinical data. On the other hand, our active learning experiments demonstrate that we can match (and even beat) ENT using only 6.6% of the training data in the clinical domain, and only 5.8% of the training data in the newswire domain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge