Anand Sampat
Composition of Experts: A Modular Compound AI System Leveraging Large Language Models
Dec 02, 2024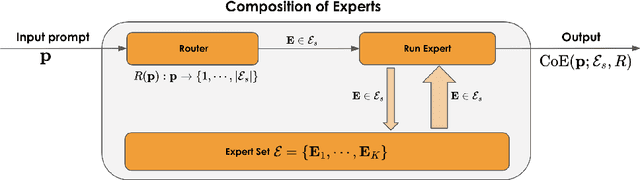
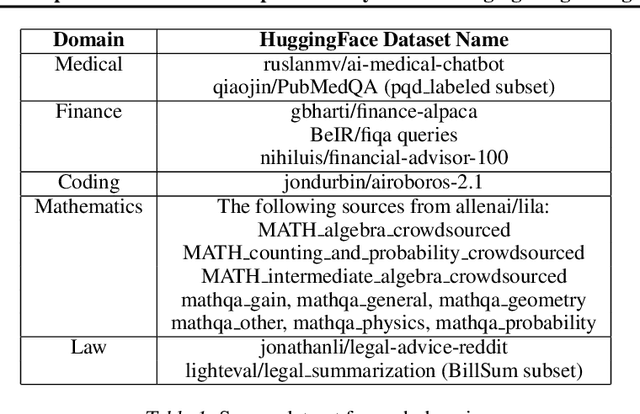
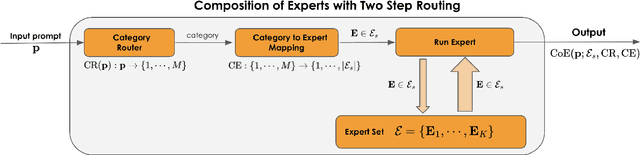
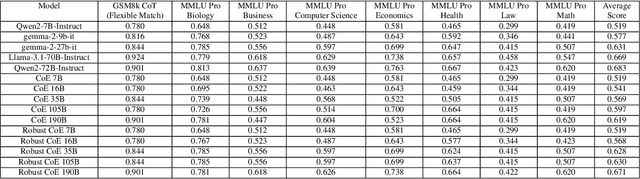
Abstract:Large Language Models (LLMs) have achieved remarkable advancements, but their monolithic nature presents challenges in terms of scalability, cost, and customization. This paper introduces the Composition of Experts (CoE), a modular compound AI system leveraging multiple expert LLMs. CoE leverages a router to dynamically select the most appropriate expert for a given input, enabling efficient utilization of resources and improved performance. We formulate the general problem of training a CoE and discuss inherent complexities associated with it. We propose a two-step routing approach to address these complexities that first uses a router to classify the input into distinct categories followed by a category-to-expert mapping to obtain desired experts. CoE offers a flexible and cost-effective solution to build compound AI systems. Our empirical evaluation demonstrates the effectiveness of CoE in achieving superior performance with reduced computational overhead. Given that CoE comprises of many expert LLMs it has unique system requirements for cost-effective serving. We present an efficient implementation of CoE leveraging SambaNova SN40L RDUs unique three-tiered memory architecture. CoEs obtained using open weight LLMs Qwen/Qwen2-7B-Instruct, google/gemma-2-9b-it, google/gemma-2-27b-it, meta-llama/Llama-3.1-70B-Instruct and Qwen/Qwen2-72B-Instruct achieve a score of $59.4$ with merely $31$ billion average active parameters on Arena-Hard and a score of $9.06$ with $54$ billion average active parameters on MT-Bench.
ContriMix: Unsupervised disentanglement of content and attribute for domain generalization in microscopy image analysis
Jul 03, 2023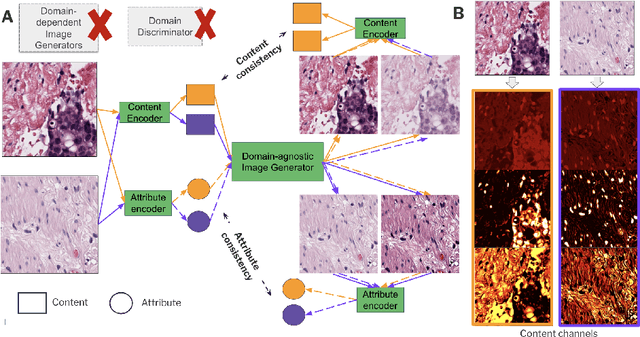
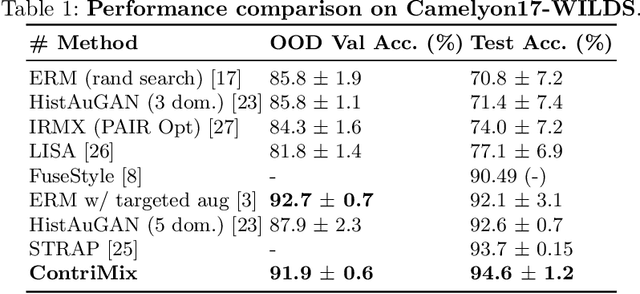
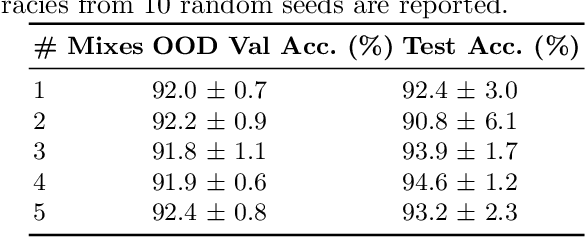
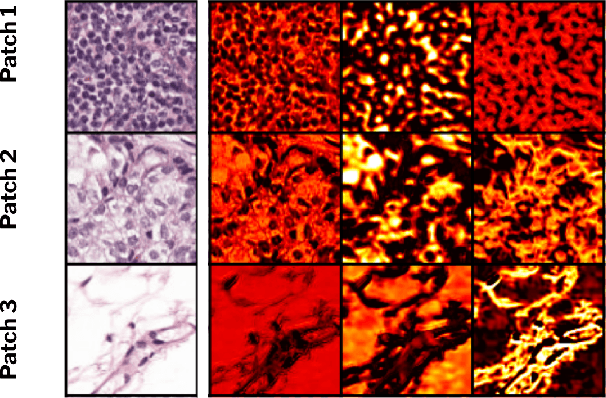
Abstract:Domain generalization is critical for real-world applications of machine learning models to microscopy images, including histopathology and fluorescence imaging. Artifacts in histopathology arise through a complex combination of factors relating to tissue collection and laboratory processing, as well as factors intrinsic to patient samples. In fluorescence imaging, these artifacts stem from variations across experimental batches. The complexity and subtlety of these artifacts make the enumeration of data domains intractable. Therefore, augmentation-based methods of domain generalization that require domain identifiers and manual fine-tuning are inadequate in this setting. To overcome this challenge, we introduce ContriMix, a domain generalization technique that learns to generate synthetic images by disentangling and permuting the biological content ("content") and technical variations ("attributes") in microscopy images. ContriMix does not rely on domain identifiers or handcrafted augmentations and makes no assumptions about the input characteristics of images. We assess the performance of ContriMix on two pathology datasets (Camelyon17-WILDS and a prostate cell classification dataset) and one fluorescence microscopy dataset (RxRx1-WILDS). ContriMix outperforms current state-of-the-art methods in all datasets, motivating its usage for microscopy image analysis in real-world settings where domain information is hard to come by.
Synthetic DOmain-Targeted Augmentation (S-DOTA) Improves Model Generalization in Digital Pathology
May 03, 2023Abstract:Machine learning algorithms have the potential to improve patient outcomes in digital pathology. However, generalization of these tools is currently limited by sensitivity to variations in tissue preparation, staining procedures and scanning equipment that lead to domain shift in digitized slides. To overcome this limitation and improve model generalization, we studied the effectiveness of two Synthetic DOmain-Targeted Augmentation (S-DOTA) methods, namely CycleGAN-enabled Scanner Transform (ST) and targeted Stain Vector Augmentation (SVA), and compared them against the International Color Consortium (ICC) profile-based color calibration (ICC Cal) method and a baseline method using traditional brightness, color and noise augmentations. We evaluated the ability of these techniques to improve model generalization to various tasks and settings: four models, two model types (tissue segmentation and cell classification), two loss functions, six labs, six scanners, and three indications (hepatocellular carcinoma (HCC), nonalcoholic steatohepatitis (NASH), prostate adenocarcinoma). We compared these methods based on the macro-averaged F1 scores on in-distribution (ID) and out-of-distribution (OOD) test sets across multiple domains, and found that S-DOTA methods (i.e., ST and SVA) led to significant improvements over ICC Cal and baseline on OOD data while maintaining comparable performance on ID data. Thus, we demonstrate that S-DOTA may help address generalization due to domain shift in real world applications.
SC-MIL: Supervised Contrastive Multiple Instance Learning for Imbalanced Classification in Pathology
Mar 23, 2023Abstract:Multiple Instance learning (MIL) models have been extensively used in pathology to predict biomarkers and risk-stratify patients from gigapixel-sized images. Machine learning problems in medical imaging often deal with rare diseases, making it important for these models to work in a label-imbalanced setting. Furthermore, these imbalances can occur in out-of-distribution (OOD) datasets when the models are deployed in the real-world. We leverage the idea that decoupling feature and classifier learning can lead to improved decision boundaries for label imbalanced datasets. To this end, we investigate the integration of supervised contrastive learning with multiple instance learning (SC-MIL). Specifically, we propose a joint-training MIL framework in the presence of label imbalance that progressively transitions from learning bag-level representations to optimal classifier learning. We perform experiments with different imbalance settings for two well-studied problems in cancer pathology: subtyping of non-small cell lung cancer and subtyping of renal cell carcinoma. SC-MIL provides large and consistent improvements over other techniques on both in-distribution (ID) and OOD held-out sets across multiple imbalanced settings.
Self-training of Machine Learning Models for Liver Histopathology: Generalization under Clinical Shifts
Nov 14, 2022

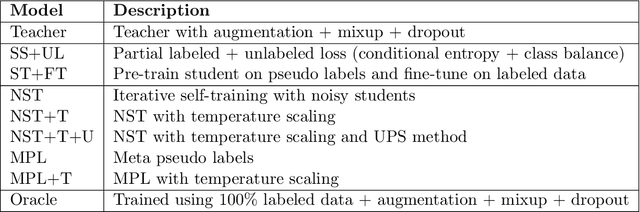

Abstract:Histopathology images are gigapixel-sized and include features and information at different resolutions. Collecting annotations in histopathology requires highly specialized pathologists, making it expensive and time-consuming. Self-training can alleviate annotation constraints by learning from both labeled and unlabeled data, reducing the amount of annotations required from pathologists. We study the design of teacher-student self-training systems for Non-alcoholic Steatohepatitis (NASH) using clinical histopathology datasets with limited annotations. We evaluate the models on in-distribution and out-of-distribution test data under clinical data shifts. We demonstrate that through self-training, the best student model statistically outperforms the teacher with a $3\%$ absolute difference on the macro F1 score. The best student model also approaches the performance of a fully supervised model trained with twice as many annotations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge