Raymond Biju
PLUTO: Pathology-Universal Transformer
May 13, 2024



Abstract:Pathology is the study of microscopic inspection of tissue, and a pathology diagnosis is often the medical gold standard to diagnose disease. Pathology images provide a unique challenge for computer-vision-based analysis: a single pathology Whole Slide Image (WSI) is gigapixel-sized and often contains hundreds of thousands to millions of objects of interest across multiple resolutions. In this work, we propose PathoLogy Universal TransfOrmer (PLUTO): a light-weight pathology FM that is pre-trained on a diverse dataset of 195 million image tiles collected from multiple sites and extracts meaningful representations across multiple WSI scales that enable a large variety of downstream pathology tasks. In particular, we design task-specific adaptation heads that utilize PLUTO's output embeddings for tasks which span pathology scales ranging from subcellular to slide-scale, including instance segmentation, tile classification, and slide-level prediction. We compare PLUTO's performance to other state-of-the-art methods on a diverse set of external and internal benchmarks covering multiple biologically relevant tasks, tissue types, resolutions, stains, and scanners. We find that PLUTO matches or outperforms existing task-specific baselines and pathology-specific foundation models, some of which use orders-of-magnitude larger datasets and model sizes when compared to PLUTO. Our findings present a path towards a universal embedding to power pathology image analysis, and motivate further exploration around pathology foundation models in terms of data diversity, architectural improvements, sample efficiency, and practical deployability in real-world applications.
Improved statistical benchmarking of digital pathology models using pairwise frames evaluation
Jun 07, 2023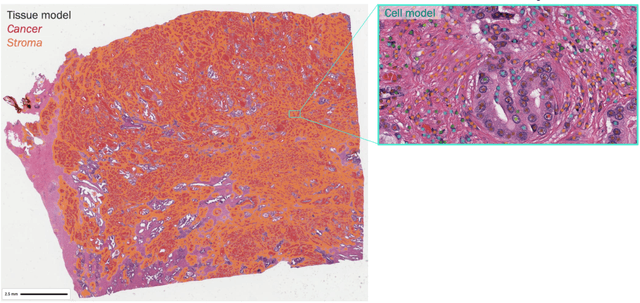
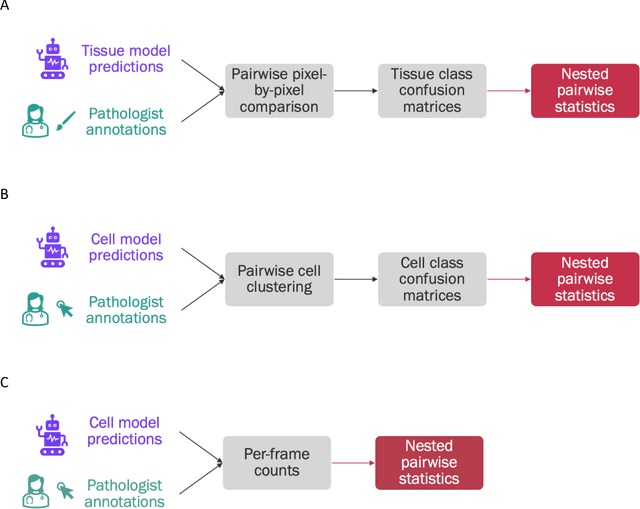
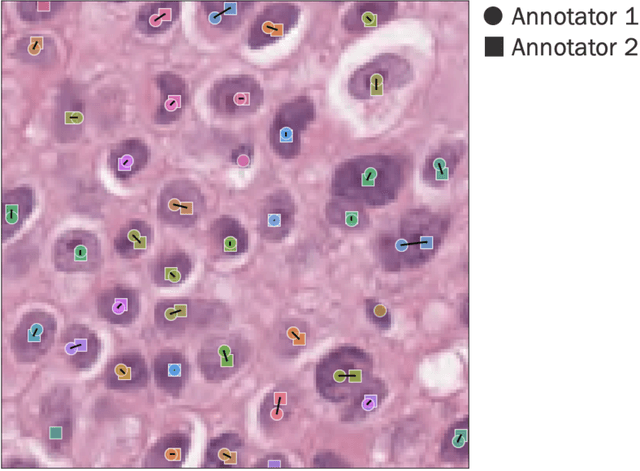
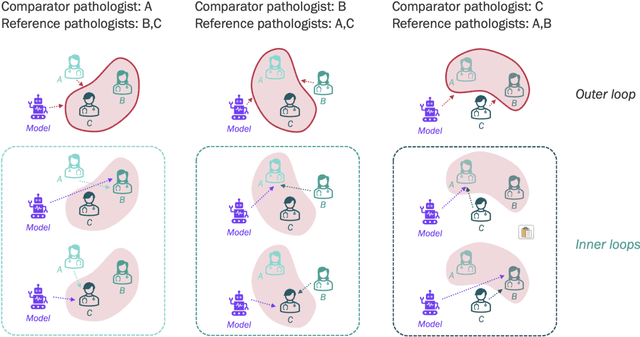
Abstract:Nested pairwise frames is a method for relative benchmarking of cell or tissue digital pathology models against manual pathologist annotations on a set of sampled patches. At a high level, the method compares agreement between a candidate model and pathologist annotations with agreement among pathologists' annotations. This evaluation framework addresses fundamental issues of data size and annotator variability in using manual pathologist annotations as a source of ground truth for model validation. We implemented nested pairwise frames evaluation for tissue classification, cell classification, and cell count prediction tasks and show results for cell and tissue models deployed on an H&E-stained melanoma dataset.
Synthetic DOmain-Targeted Augmentation (S-DOTA) Improves Model Generalization in Digital Pathology
May 03, 2023Abstract:Machine learning algorithms have the potential to improve patient outcomes in digital pathology. However, generalization of these tools is currently limited by sensitivity to variations in tissue preparation, staining procedures and scanning equipment that lead to domain shift in digitized slides. To overcome this limitation and improve model generalization, we studied the effectiveness of two Synthetic DOmain-Targeted Augmentation (S-DOTA) methods, namely CycleGAN-enabled Scanner Transform (ST) and targeted Stain Vector Augmentation (SVA), and compared them against the International Color Consortium (ICC) profile-based color calibration (ICC Cal) method and a baseline method using traditional brightness, color and noise augmentations. We evaluated the ability of these techniques to improve model generalization to various tasks and settings: four models, two model types (tissue segmentation and cell classification), two loss functions, six labs, six scanners, and three indications (hepatocellular carcinoma (HCC), nonalcoholic steatohepatitis (NASH), prostate adenocarcinoma). We compared these methods based on the macro-averaged F1 scores on in-distribution (ID) and out-of-distribution (OOD) test sets across multiple domains, and found that S-DOTA methods (i.e., ST and SVA) led to significant improvements over ICC Cal and baseline on OOD data while maintaining comparable performance on ID data. Thus, we demonstrate that S-DOTA may help address generalization due to domain shift in real world applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge