Aaditya Prakash
ContriMix: Unsupervised disentanglement of content and attribute for domain generalization in microscopy image analysis
Jul 03, 2023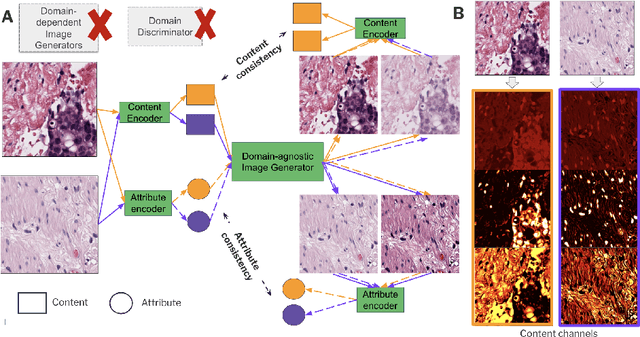
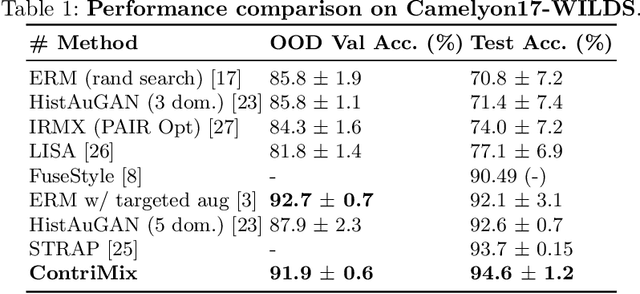
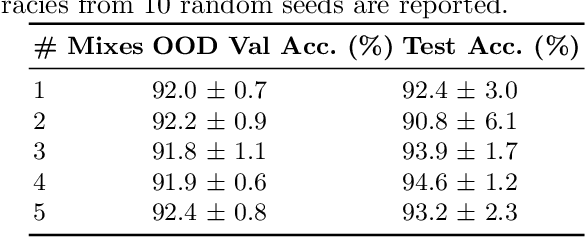
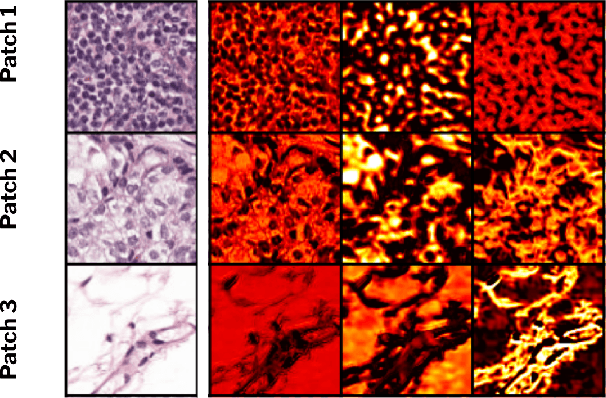
Abstract:Domain generalization is critical for real-world applications of machine learning models to microscopy images, including histopathology and fluorescence imaging. Artifacts in histopathology arise through a complex combination of factors relating to tissue collection and laboratory processing, as well as factors intrinsic to patient samples. In fluorescence imaging, these artifacts stem from variations across experimental batches. The complexity and subtlety of these artifacts make the enumeration of data domains intractable. Therefore, augmentation-based methods of domain generalization that require domain identifiers and manual fine-tuning are inadequate in this setting. To overcome this challenge, we introduce ContriMix, a domain generalization technique that learns to generate synthetic images by disentangling and permuting the biological content ("content") and technical variations ("attributes") in microscopy images. ContriMix does not rely on domain identifiers or handcrafted augmentations and makes no assumptions about the input characteristics of images. We assess the performance of ContriMix on two pathology datasets (Camelyon17-WILDS and a prostate cell classification dataset) and one fluorescence microscopy dataset (RxRx1-WILDS). ContriMix outperforms current state-of-the-art methods in all datasets, motivating its usage for microscopy image analysis in real-world settings where domain information is hard to come by.
Synthetic DOmain-Targeted Augmentation (S-DOTA) Improves Model Generalization in Digital Pathology
May 03, 2023Abstract:Machine learning algorithms have the potential to improve patient outcomes in digital pathology. However, generalization of these tools is currently limited by sensitivity to variations in tissue preparation, staining procedures and scanning equipment that lead to domain shift in digitized slides. To overcome this limitation and improve model generalization, we studied the effectiveness of two Synthetic DOmain-Targeted Augmentation (S-DOTA) methods, namely CycleGAN-enabled Scanner Transform (ST) and targeted Stain Vector Augmentation (SVA), and compared them against the International Color Consortium (ICC) profile-based color calibration (ICC Cal) method and a baseline method using traditional brightness, color and noise augmentations. We evaluated the ability of these techniques to improve model generalization to various tasks and settings: four models, two model types (tissue segmentation and cell classification), two loss functions, six labs, six scanners, and three indications (hepatocellular carcinoma (HCC), nonalcoholic steatohepatitis (NASH), prostate adenocarcinoma). We compared these methods based on the macro-averaged F1 scores on in-distribution (ID) and out-of-distribution (OOD) test sets across multiple domains, and found that S-DOTA methods (i.e., ST and SVA) led to significant improvements over ICC Cal and baseline on OOD data while maintaining comparable performance on ID data. Thus, we demonstrate that S-DOTA may help address generalization due to domain shift in real world applications.
Additive MIL: Intrinsic Interpretability for Pathology
Jun 03, 2022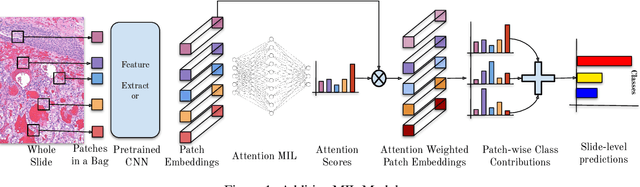
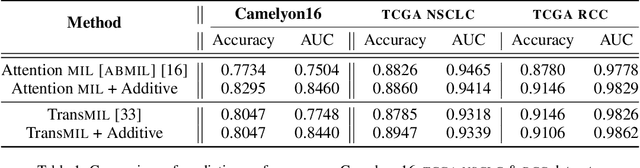
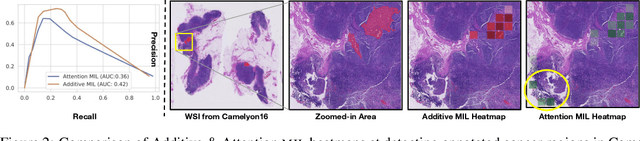

Abstract:Multiple Instance Learning (MIL) has been widely applied in pathology towards solving critical problems such as automating cancer diagnosis and grading, predicting patient prognosis, and therapy response. Deploying these models in a clinical setting requires careful inspection of these black boxes during development and deployment to identify failures and maintain physician trust. In this work, we propose a simple formulation of MIL models, which enables interpretability while maintaining similar predictive performance. Our Additive MIL models enable spatial credit assignment such that the contribution of each region in the image can be exactly computed and visualized. We show that our spatial credit assignment coincides with regions used by pathologists during diagnosis and improves upon classical attention heatmaps from attention MIL models. We show that any existing MIL model can be made additive with a simple change in function composition. We also show how these models can debug model failures, identify spurious features, and highlight class-wise regions of interest, enabling their use in high-stakes environments such as clinical decision-making.
Rethinking Machine Learning Model Evaluation in Pathology
Apr 18, 2022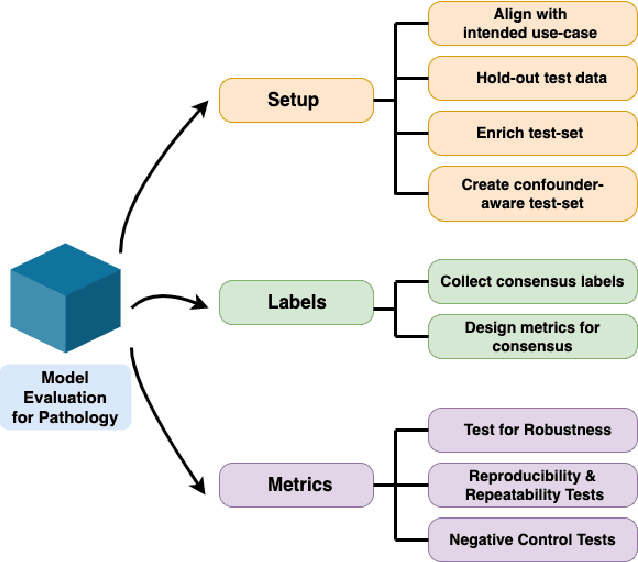
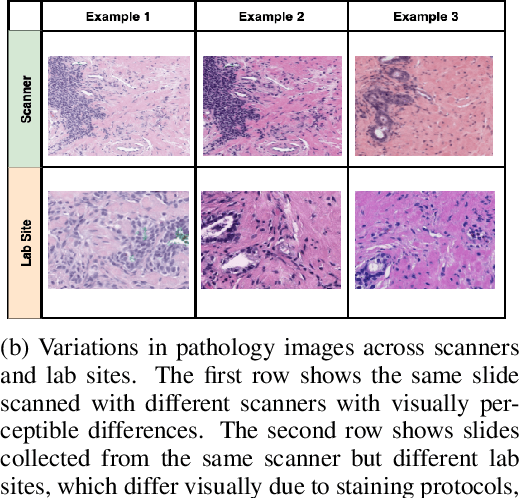
Abstract:Machine Learning has been applied to pathology images in research and clinical practice with promising outcomes. However, standard ML models often lack the rigorous evaluation required for clinical decisions. Machine learning techniques for natural images are ill-equipped to deal with pathology images that are significantly large and noisy, require expensive labeling, are hard to interpret, and are susceptible to spurious correlations. We propose a set of practical guidelines for ML evaluation in pathology that address the above concerns. The paper includes measures for setting up the evaluation framework, effectively dealing with variability in labels, and a recommended suite of tests to address issues related to domain shift, robustness, and confounding variables. We hope that the proposed framework will bridge the gap between ML researchers and domain experts, leading to wider adoption of ML techniques in pathology and improving patient outcomes.
RePr: Improved Training of Convolutional Filters
Nov 26, 2018



Abstract:A well-trained Convolutional Neural Network can easily be pruned without significant loss of performance. This is because of unnecessary overlap in the features captured by the network's filters. Innovations in network architecture such as skip/dense connections and Inception units have mitigated this problem to some extent, but these improvements come with increased computation and memory requirements at run-time. We attempt to address this problem from another angle - not by changing the network structure but by altering the training method. We show that by temporarily pruning and then restoring a subset of the model's filters, and repeating this process cyclically, overlap in the learned features is reduced, producing improved generalization. We show that the existing model-pruning criteria are not optimal for selecting filters to prune in this context and introduce inter-filter orthogonality as the ranking criteria to determine under-expressive filters. Our method is applicable both to vanilla convolutional networks and more complex modern architectures, and improves the performance across a variety of tasks, especially when applied to smaller networks.
DR-BiLSTM: Dependent Reading Bidirectional LSTM for Natural Language Inference
Apr 11, 2018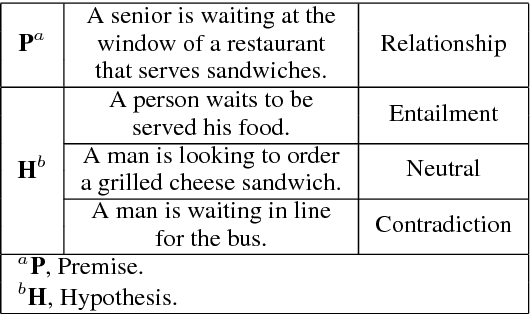
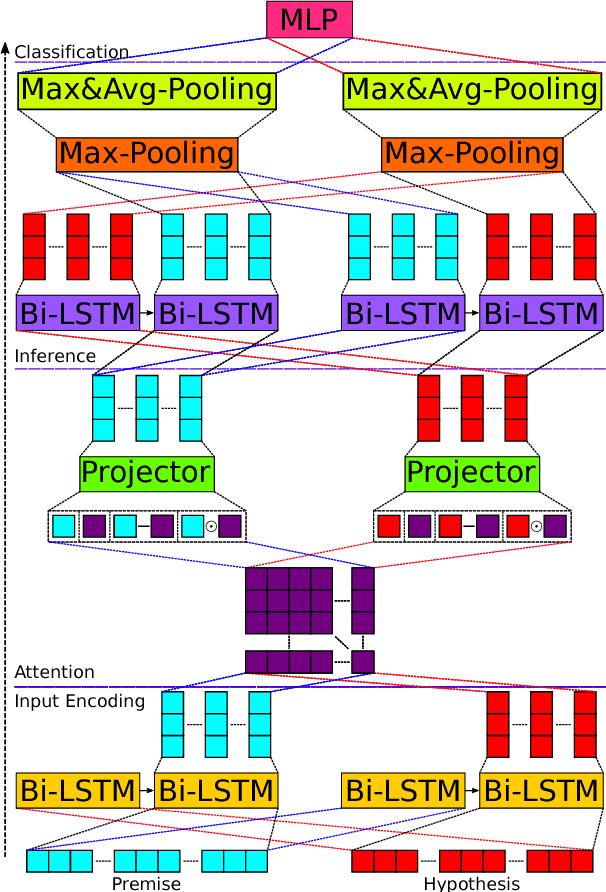
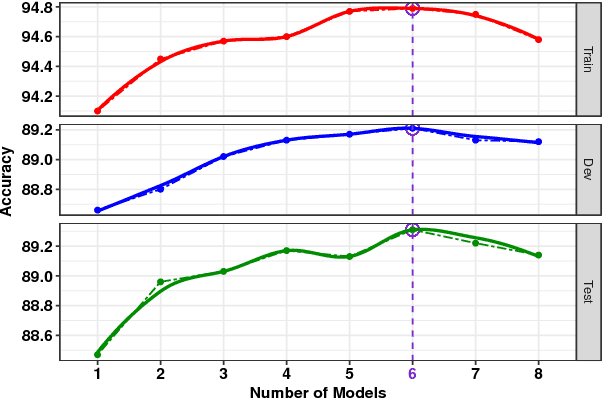
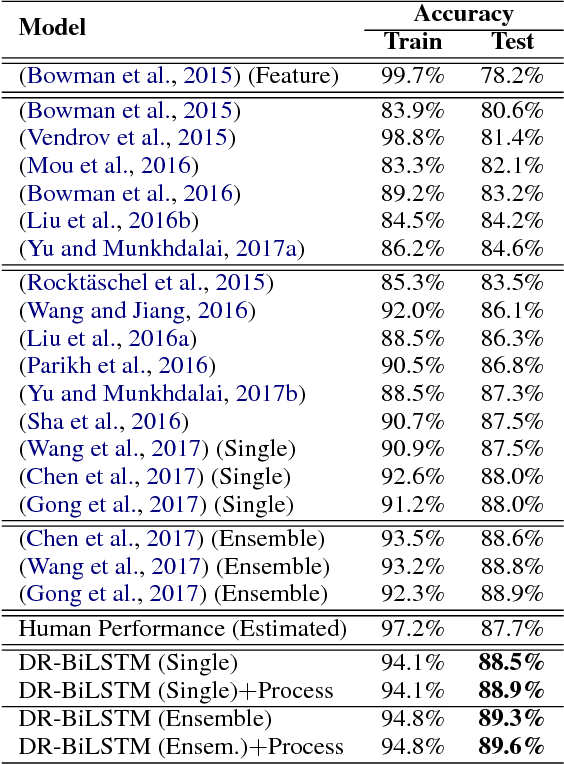
Abstract:We present a novel deep learning architecture to address the natural language inference (NLI) task. Existing approaches mostly rely on simple reading mechanisms for independent encoding of the premise and hypothesis. Instead, we propose a novel dependent reading bidirectional LSTM network (DR-BiLSTM) to efficiently model the relationship between a premise and a hypothesis during encoding and inference. We also introduce a sophisticated ensemble strategy to combine our proposed models, which noticeably improves final predictions. Finally, we demonstrate how the results can be improved further with an additional preprocessing step. Our evaluation shows that DR-BiLSTM obtains the best single model and ensemble model results achieving the new state-of-the-art scores on the Stanford NLI dataset.
Deflecting Adversarial Attacks with Pixel Deflection
Mar 30, 2018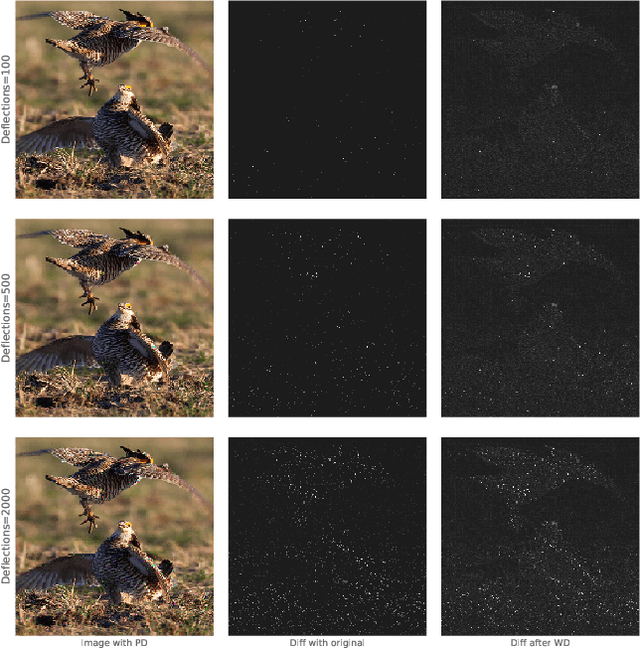
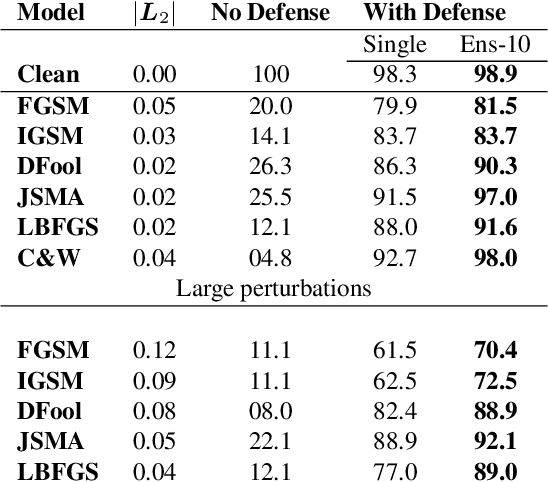
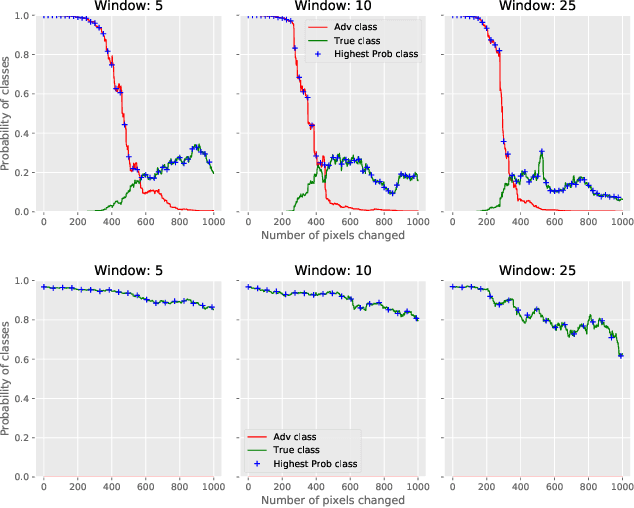
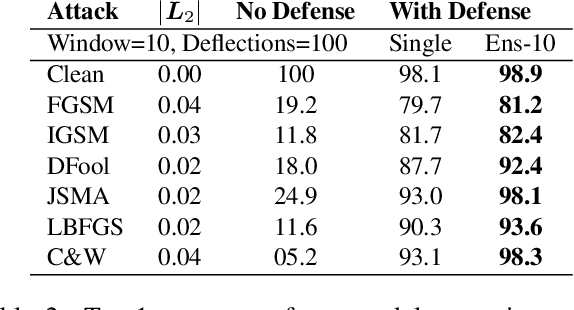
Abstract:CNNs are poised to become integral parts of many critical systems. Despite their robustness to natural variations, image pixel values can be manipulated, via small, carefully crafted, imperceptible perturbations, to cause a model to misclassify images. We present an algorithm to process an image so that classification accuracy is significantly preserved in the presence of such adversarial manipulations. Image classifiers tend to be robust to natural noise, and adversarial attacks tend to be agnostic to object location. These observations motivate our strategy, which leverages model robustness to defend against adversarial perturbations by forcing the image to match natural image statistics. Our algorithm locally corrupts the image by redistributing pixel values via a process we term pixel deflection. A subsequent wavelet-based denoising operation softens this corruption, as well as some of the adversarial changes. We demonstrate experimentally that the combination of these techniques enables the effective recovery of the true class, against a variety of robust attacks. Our results compare favorably with current state-of-the-art defenses, without requiring retraining or modifying the CNN.
Protecting JPEG Images Against Adversarial Attacks
Mar 02, 2018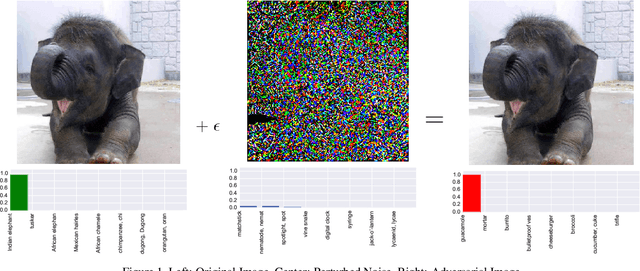
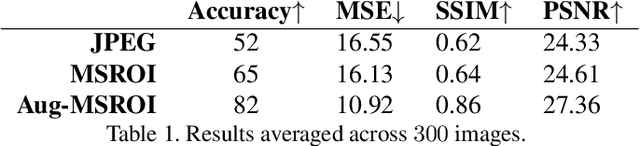
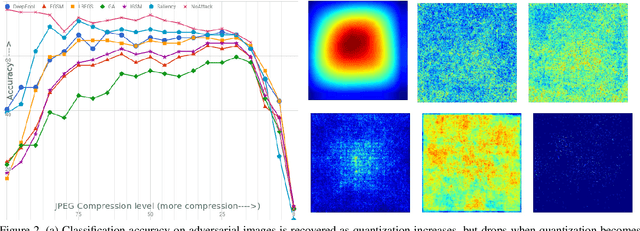
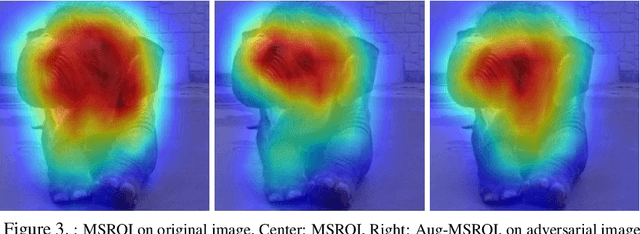
Abstract:As deep neural networks (DNNs) have been integrated into critical systems, several methods to attack these systems have been developed. These adversarial attacks make imperceptible modifications to an image that fool DNN classifiers. We present an adaptive JPEG encoder which defends against many of these attacks. Experimentally, we show that our method produces images with high visual quality while greatly reducing the potency of state-of-the-art attacks. Our algorithm requires only a modest increase in encoding time, produces a compressed image which can be decompressed by an off-the-shelf JPEG decoder, and classified by an unmodified classifier
Semantic Perceptual Image Compression using Deep Convolution Networks
Mar 29, 2017
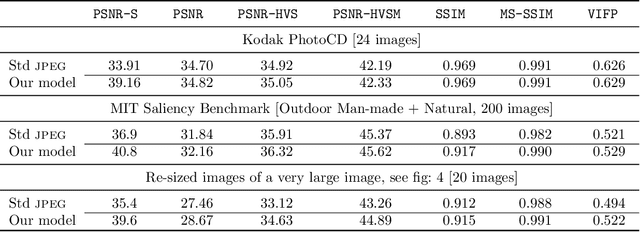


Abstract:It has long been considered a significant problem to improve the visual quality of lossy image and video compression. Recent advances in computing power together with the availability of large training data sets has increased interest in the application of deep learning cnns to address image recognition and image processing tasks. Here, we present a powerful cnn tailored to the specific task of semantic image understanding to achieve higher visual quality in lossy compression. A modest increase in complexity is incorporated to the encoder which allows a standard, off-the-shelf jpeg decoder to be used. While jpeg encoding may be optimized for generic images, the process is ultimately unaware of the specific content of the image to be compressed. Our technique makes jpeg content-aware by designing and training a model to identify multiple semantic regions in a given image. Unlike object detection techniques, our model does not require labeling of object positions and is able to identify objects in a single pass. We present a new cnn architecture directed specifically to image compression, which generates a map that highlights semantically-salient regions so that they can be encoded at higher quality as compared to background regions. By adding a complete set of features for every class, and then taking a threshold over the sum of all feature activations, we generate a map that highlights semantically-salient regions so that they can be encoded at a better quality compared to background regions. Experiments are presented on the Kodak PhotoCD dataset and the MIT Saliency Benchmark dataset, in which our algorithm achieves higher visual quality for the same compressed size.
Condensed Memory Networks for Clinical Diagnostic Inferencing
Jan 03, 2017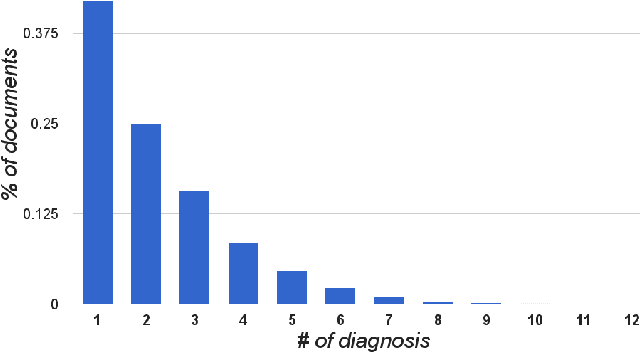
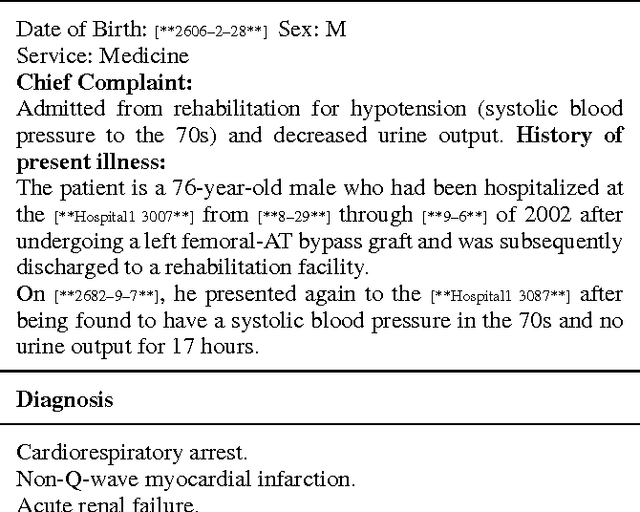
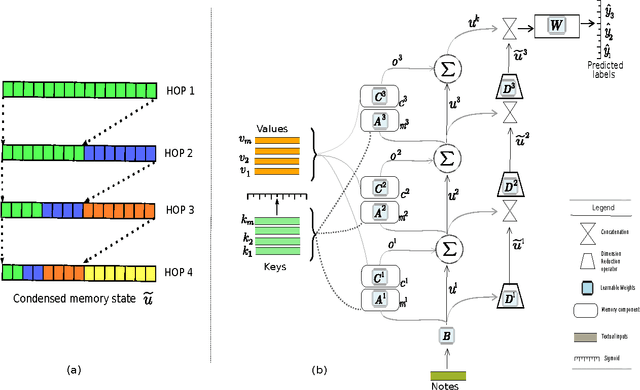
Abstract:Diagnosis of a clinical condition is a challenging task, which often requires significant medical investigation. Previous work related to diagnostic inferencing problems mostly consider multivariate observational data (e.g. physiological signals, lab tests etc.). In contrast, we explore the problem using free-text medical notes recorded in an electronic health record (EHR). Complex tasks like these can benefit from structured knowledge bases, but those are not scalable. We instead exploit raw text from Wikipedia as a knowledge source. Memory networks have been demonstrated to be effective in tasks which require comprehension of free-form text. They use the final iteration of the learned representation to predict probable classes. We introduce condensed memory neural networks (C-MemNNs), a novel model with iterative condensation of memory representations that preserves the hierarchy of features in the memory. Experiments on the MIMIC-III dataset show that the proposed model outperforms other variants of memory networks to predict the most probable diagnoses given a complex clinical scenario.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge