Sadid A. Hasan
SemClinBr -- a multi institutional and multi specialty semantically annotated corpus for Portuguese clinical NLP tasks
Jan 27, 2020



Abstract:The high volume of research focusing on extracting patient's information from electronic health records (EHR) has led to an increase in the demand for annotated corpora, which are a very valuable resource for both the development and evaluation of natural language processing (NLP) algorithms. The absence of a multi-purpose clinical corpus outside the scope of the English language, especially in Brazilian Portuguese, is glaring and severely impacts scientific progress in the biomedical NLP field. In this study, we developed a semantically annotated corpus using clinical texts from multiple medical specialties, document types, and institutions. We present the following: (1) a survey listing common aspects and lessons learned from previous research, (2) a fine-grained annotation schema which could be replicated and guide other annotation initiatives, (3) a web-based annotation tool focusing on an annotation suggestion feature, and (4) both intrinsic and extrinsic evaluation of the annotations. The result of this work is the SemClinBr, a corpus that has 1,000 clinical notes, labeled with 65,117 entities and 11,263 relations, and can support a variety of clinical NLP tasks and boost the EHR's secondary use for the Portuguese language.
DR-BiLSTM: Dependent Reading Bidirectional LSTM for Natural Language Inference
Apr 11, 2018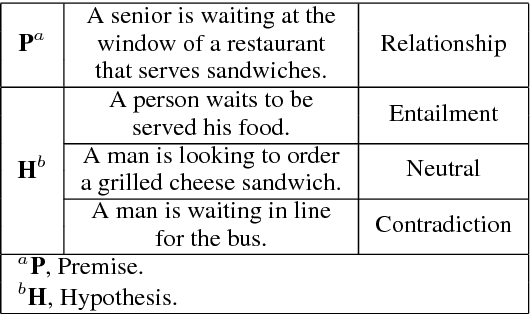
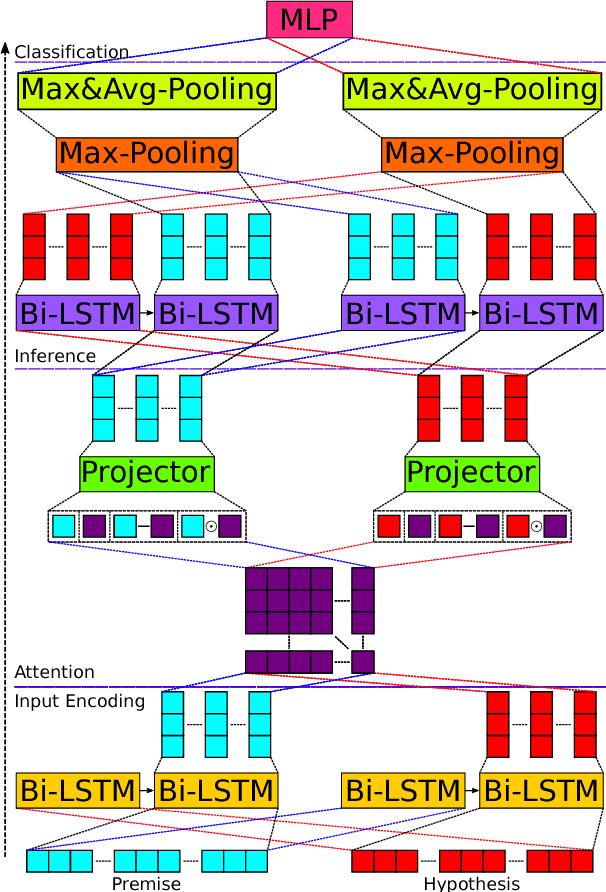
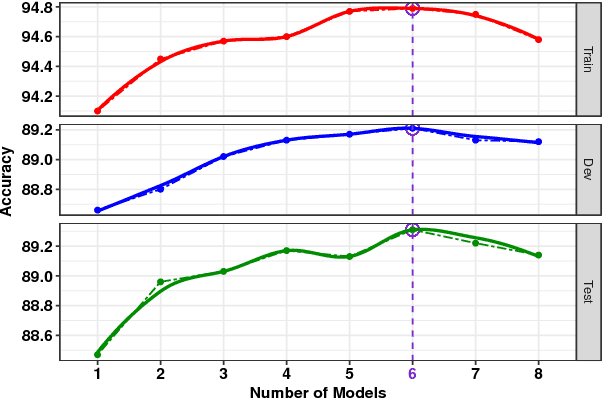
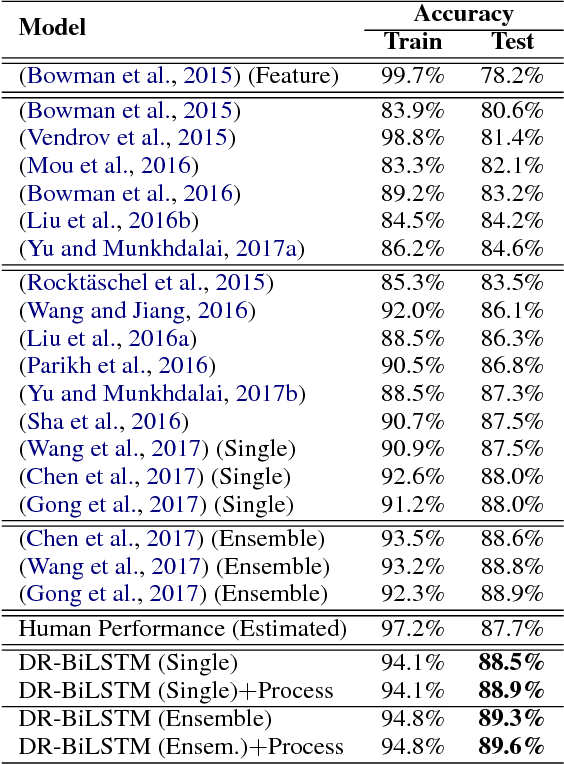
Abstract:We present a novel deep learning architecture to address the natural language inference (NLI) task. Existing approaches mostly rely on simple reading mechanisms for independent encoding of the premise and hypothesis. Instead, we propose a novel dependent reading bidirectional LSTM network (DR-BiLSTM) to efficiently model the relationship between a premise and a hypothesis during encoding and inference. We also introduce a sophisticated ensemble strategy to combine our proposed models, which noticeably improves final predictions. Finally, we demonstrate how the results can be improved further with an additional preprocessing step. Our evaluation shows that DR-BiLSTM obtains the best single model and ensemble model results achieving the new state-of-the-art scores on the Stanford NLI dataset.
Condensed Memory Networks for Clinical Diagnostic Inferencing
Jan 03, 2017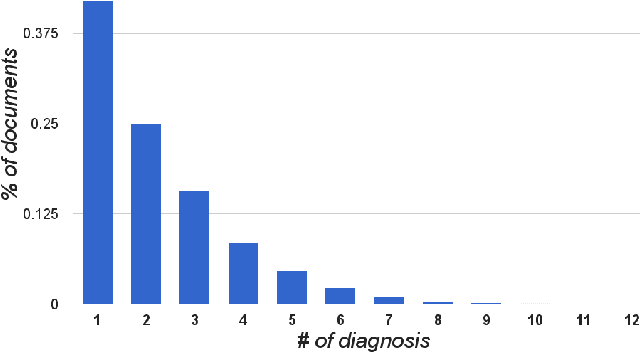
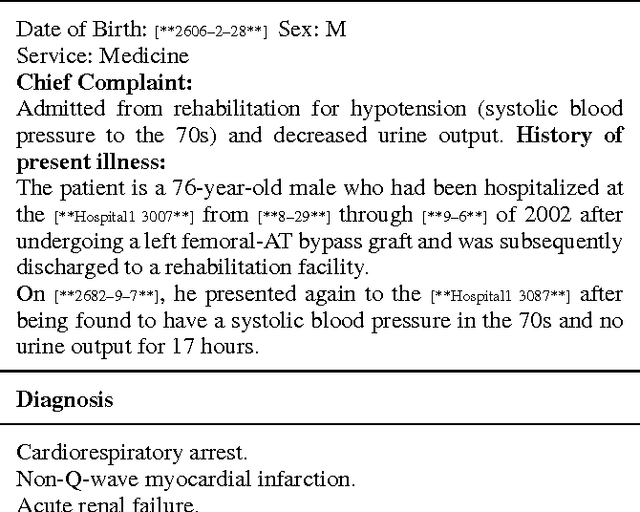
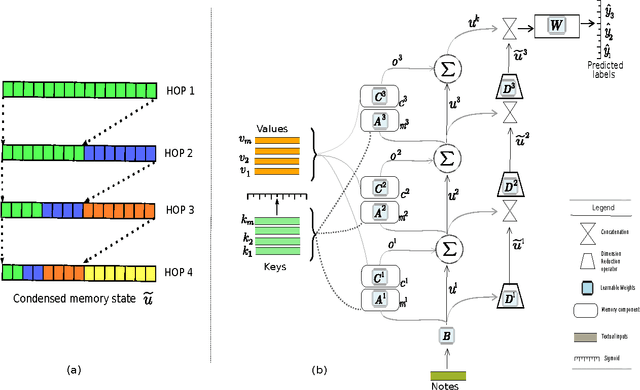
Abstract:Diagnosis of a clinical condition is a challenging task, which often requires significant medical investigation. Previous work related to diagnostic inferencing problems mostly consider multivariate observational data (e.g. physiological signals, lab tests etc.). In contrast, we explore the problem using free-text medical notes recorded in an electronic health record (EHR). Complex tasks like these can benefit from structured knowledge bases, but those are not scalable. We instead exploit raw text from Wikipedia as a knowledge source. Memory networks have been demonstrated to be effective in tasks which require comprehension of free-form text. They use the final iteration of the learned representation to predict probable classes. We introduce condensed memory neural networks (C-MemNNs), a novel model with iterative condensation of memory representations that preserves the hierarchy of features in the memory. Experiments on the MIMIC-III dataset show that the proposed model outperforms other variants of memory networks to predict the most probable diagnoses given a complex clinical scenario.
Neural Paraphrase Generation with Stacked Residual LSTM Networks
Oct 13, 2016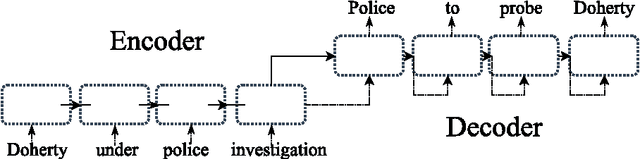

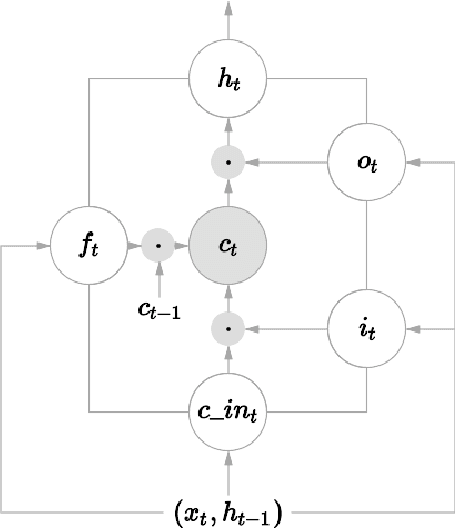
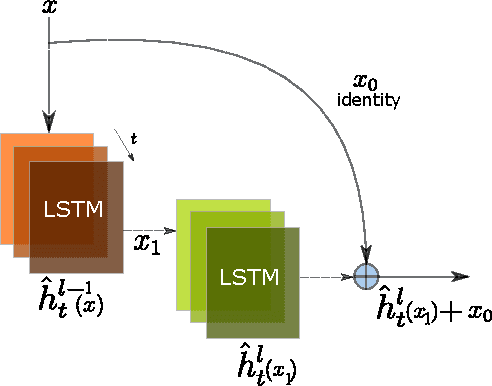
Abstract:In this paper, we propose a novel neural approach for paraphrase generation. Conventional para- phrase generation methods either leverage hand-written rules and thesauri-based alignments, or use statistical machine learning principles. To the best of our knowledge, this work is the first to explore deep learning models for paraphrase generation. Our primary contribution is a stacked residual LSTM network, where we add residual connections between LSTM layers. This allows for efficient training of deep LSTMs. We evaluate our model and other state-of-the-art deep learning models on three different datasets: PPDB, WikiAnswers and MSCOCO. Evaluation results demonstrate that our model outperforms sequence to sequence, attention-based and bi- directional LSTM models on BLEU, METEOR, TER and an embedding-based sentence similarity metric.
Complex Question Answering: Unsupervised Learning Approaches and Experiments
Jan 15, 2014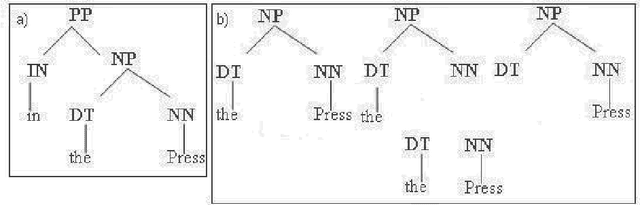
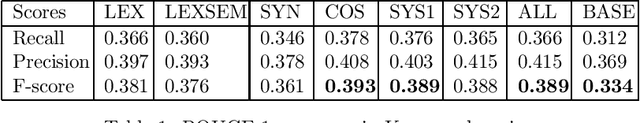
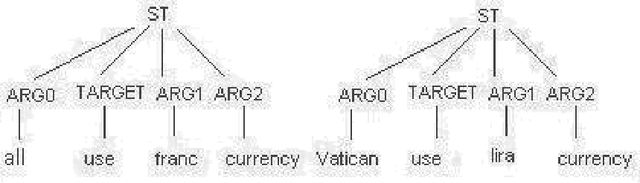
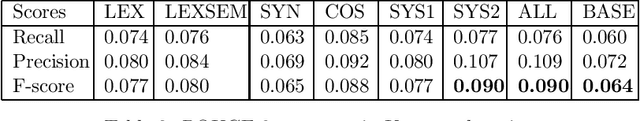
Abstract:Complex questions that require inferencing and synthesizing information from multiple documents can be seen as a kind of topic-oriented, informative multi-document summarization where the goal is to produce a single text as a compressed version of a set of documents with a minimum loss of relevant information. In this paper, we experiment with one empirical method and two unsupervised statistical machine learning techniques: K-means and Expectation Maximization (EM), for computing relative importance of the sentences. We compare the results of these approaches. Our experiments show that the empirical approach outperforms the other two techniques and EM performs better than K-means. However, the performance of these approaches depends entirely on the feature set used and the weighting of these features. In order to measure the importance and relevance to the user query we extract different kinds of features (i.e. lexical, lexical semantic, cosine similarity, basic element, tree kernel based syntactic and shallow-semantic) for each of the document sentences. We use a local search technique to learn the weights of the features. To the best of our knowledge, no study has used tree kernel functions to encode syntactic/semantic information for more complex tasks such as computing the relatedness between the query sentences and the document sentences in order to generate query-focused summaries (or answers to complex questions). For each of our methods of generating summaries (i.e. empirical, K-means and EM) we show the effects of syntactic and shallow-semantic features over the bag-of-words (BOW) features.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge