Siddhant Shingi
SC-MIL: Supervised Contrastive Multiple Instance Learning for Imbalanced Classification in Pathology
Mar 23, 2023Abstract:Multiple Instance learning (MIL) models have been extensively used in pathology to predict biomarkers and risk-stratify patients from gigapixel-sized images. Machine learning problems in medical imaging often deal with rare diseases, making it important for these models to work in a label-imbalanced setting. Furthermore, these imbalances can occur in out-of-distribution (OOD) datasets when the models are deployed in the real-world. We leverage the idea that decoupling feature and classifier learning can lead to improved decision boundaries for label imbalanced datasets. To this end, we investigate the integration of supervised contrastive learning with multiple instance learning (SC-MIL). Specifically, we propose a joint-training MIL framework in the presence of label imbalance that progressively transitions from learning bag-level representations to optimal classifier learning. We perform experiments with different imbalance settings for two well-studied problems in cancer pathology: subtyping of non-small cell lung cancer and subtyping of renal cell carcinoma. SC-MIL provides large and consistent improvements over other techniques on both in-distribution (ID) and OOD held-out sets across multiple imbalanced settings.
Interpretability of Epidemiological Models : The Curse of Non-Identifiability
Apr 30, 2021
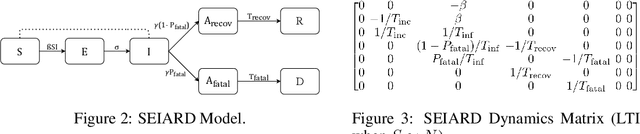
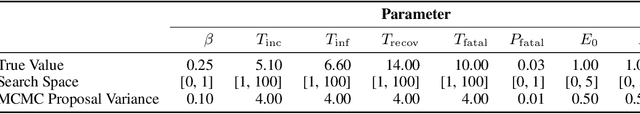

Abstract:Interpretability of epidemiological models is a key consideration, especially when these models are used in a public health setting. Interpretability is strongly linked to the identifiability of the underlying model parameters, i.e., the ability to estimate parameter values with high confidence given observations. In this paper, we define three separate notions of identifiability that explore the different roles played by the model definition, the loss function, the fitting methodology, and the quality and quantity of data. We define an epidemiological compartmental model framework in which we highlight these non-identifiability issues and their mitigation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge