David Page
University of Wisconsin Madison
Training Video Foundation Models with NVIDIA NeMo
Mar 17, 2025Abstract:Video Foundation Models (VFMs) have recently been used to simulate the real world to train physical AI systems and develop creative visual experiences. However, there are significant challenges in training large-scale, high quality VFMs that can generate high-quality videos. We present a scalable, open-source VFM training pipeline with NVIDIA NeMo, providing accelerated video dataset curation, multimodal data loading, and parallelized video diffusion model training and inference. We also provide a comprehensive performance analysis highlighting best practices for efficient VFM training and inference.
Cosmos World Foundation Model Platform for Physical AI
Jan 07, 2025



Abstract:Physical AI needs to be trained digitally first. It needs a digital twin of itself, the policy model, and a digital twin of the world, the world model. In this paper, we present the Cosmos World Foundation Model Platform to help developers build customized world models for their Physical AI setups. We position a world foundation model as a general-purpose world model that can be fine-tuned into customized world models for downstream applications. Our platform covers a video curation pipeline, pre-trained world foundation models, examples of post-training of pre-trained world foundation models, and video tokenizers. To help Physical AI builders solve the most critical problems of our society, we make our platform open-source and our models open-weight with permissive licenses available via https://github.com/NVIDIA/Cosmos.
MALADE: Orchestration of LLM-powered Agents with Retrieval Augmented Generation for Pharmacovigilance
Aug 03, 2024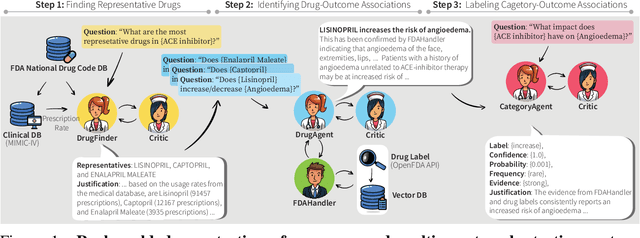
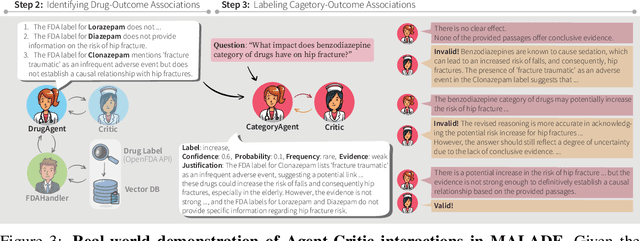
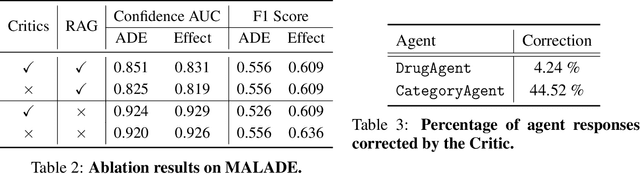
Abstract:In the era of Large Language Models (LLMs), given their remarkable text understanding and generation abilities, there is an unprecedented opportunity to develop new, LLM-based methods for trustworthy medical knowledge synthesis, extraction and summarization. This paper focuses on the problem of Pharmacovigilance (PhV), where the significance and challenges lie in identifying Adverse Drug Events (ADEs) from diverse text sources, such as medical literature, clinical notes, and drug labels. Unfortunately, this task is hindered by factors including variations in the terminologies of drugs and outcomes, and ADE descriptions often being buried in large amounts of narrative text. We present MALADE, the first effective collaborative multi-agent system powered by LLM with Retrieval Augmented Generation for ADE extraction from drug label data. This technique involves augmenting a query to an LLM with relevant information extracted from text resources, and instructing the LLM to compose a response consistent with the augmented data. MALADE is a general LLM-agnostic architecture, and its unique capabilities are: (1) leveraging a variety of external sources, such as medical literature, drug labels, and FDA tools (e.g., OpenFDA drug information API), (2) extracting drug-outcome association in a structured format along with the strength of the association, and (3) providing explanations for established associations. Instantiated with GPT-4 Turbo or GPT-4o, and FDA drug label data, MALADE demonstrates its efficacy with an Area Under ROC Curve of 0.90 against the OMOP Ground Truth table of ADEs. Our implementation leverages the Langroid multi-agent LLM framework and can be found at https://github.com/jihyechoi77/malade.
Neural Markov Prolog
Nov 27, 2023Abstract:The recent rapid advance of AI has been driven largely by innovations in neural network architectures. A concomitant concern is how to understand these resulting systems. In this paper, we propose a tool to assist in both the design of further innovative architectures and the simple yet precise communication of their structure. We propose the language Neural Markov Prolog (NMP), based on both Markov logic and Prolog, as a means to both bridge first order logic and neural network design and to allow for the easy generation and presentation of architectures for images, text, relational databases, or other target data types or their mixtures.
Differentially Private Multi-Site Treatment Effect Estimation
Oct 10, 2023Abstract:Patient privacy is a major barrier to healthcare AI. For confidentiality reasons, most patient data remains in silo in separate hospitals, preventing the design of data-driven healthcare AI systems that need large volumes of patient data to make effective decisions. A solution to this is collective learning across multiple sites through federated learning with differential privacy. However, literature in this space typically focuses on differentially private statistical estimation and machine learning, which is different from the causal inference-related problems that arise in healthcare. In this work, we take a fresh look at federated learning with a focus on causal inference; specifically, we look at estimating the average treatment effect (ATE), an important task in causal inference for healthcare applications, and provide a federated analytics approach to enable ATE estimation across multiple sites along with differential privacy (DP) guarantees at each site. The main challenge comes from site heterogeneity -- different sites have different sample sizes and privacy budgets. We address this through a class of per-site estimation algorithms that reports the ATE estimate and its variance as a quality measure, and an aggregation algorithm on the server side that minimizes the overall variance of the final ATE estimate. Our experiments on real and synthetic data show that our method reliably aggregates private statistics across sites and provides better privacy-utility tradeoff under site heterogeneity than baselines.
On Neural Networks as Infinite Tree-Structured Probabilistic Graphical Models
May 27, 2023Abstract:Deep neural networks (DNNs) lack the precise semantics and definitive probabilistic interpretation of probabilistic graphical models (PGMs). In this paper, we propose an innovative solution by constructing infinite tree-structured PGMs that correspond exactly to neural networks. Our research reveals that DNNs, during forward propagation, indeed perform approximations of PGM inference that are precise in this alternative PGM structure. Not only does our research complement existing studies that describe neural networks as kernel machines or infinite-sized Gaussian processes, it also elucidates a more direct approximation that DNNs make to exact inference in PGMs. Potential benefits include improved pedagogy and interpretation of DNNs, and algorithms that can merge the strengths of PGMs and DNNs.
From Feature Importance to Distance Metric: An Almost Exact Matching Approach for Causal Inference
Feb 23, 2023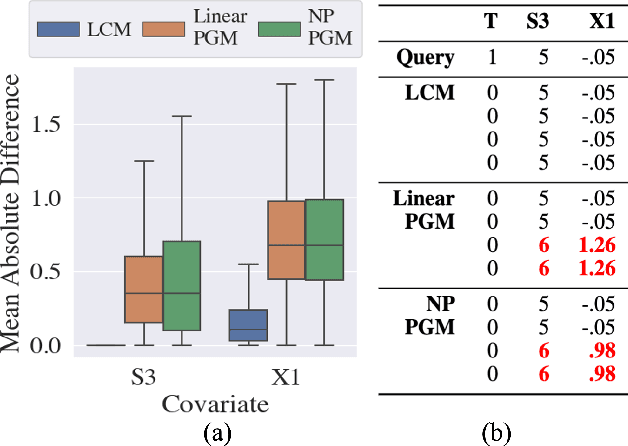
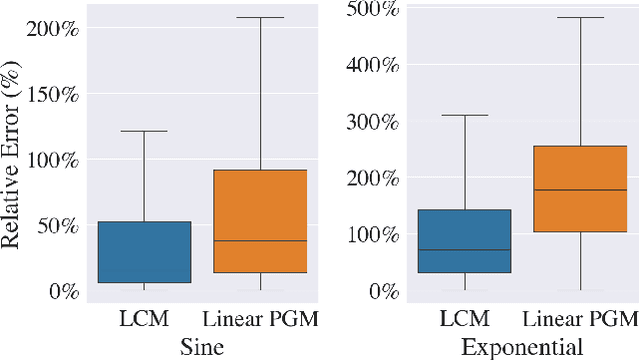
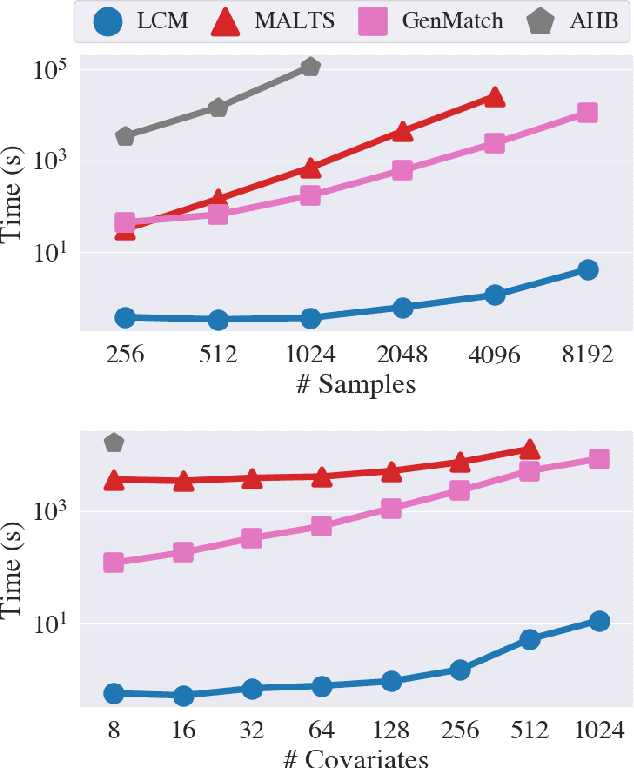
Abstract:Our goal is to produce methods for observational causal inference that are auditable, easy to troubleshoot, yield accurate treatment effect estimates, and scalable to high-dimensional data. We describe an almost-exact matching approach that achieves these goals by (i) learning a distance metric via outcome modeling, (ii) creating matched groups using the distance metric, and (iii) using the matched groups to estimate treatment effects. Our proposed method uses variable importance measurements to construct a distance metric, making it a flexible method that can be adapted to various applications. Concentrating on the scalability of the problem in the number of potential confounders, we operationalize our approach with LASSO. We derive performance guarantees for settings where LASSO outcome modeling consistently identifies all confounders (importantly without requiring the linear model to be correctly specified). We also provide experimental results demonstrating the auditability of matches, as well as extensions to more general nonparametric outcome modeling.
Predicting Drug-Drug Interactions from Heterogeneous Data: An Embedding Approach
Mar 19, 2021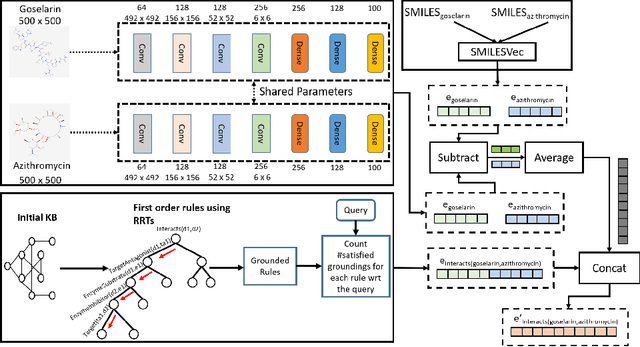
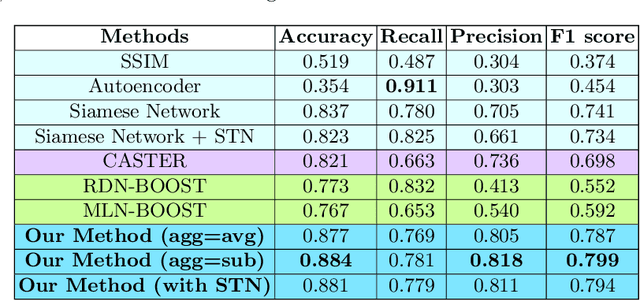
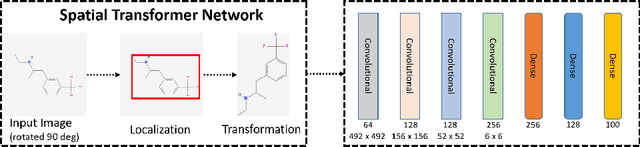
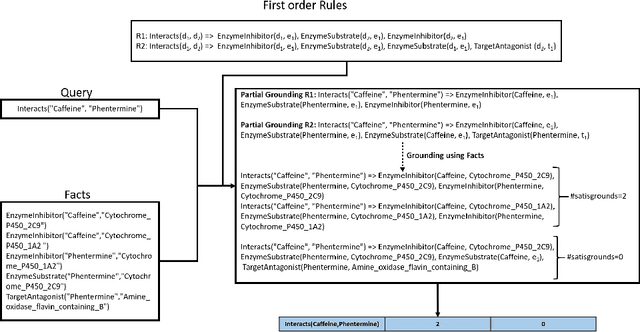
Abstract:Predicting and discovering drug-drug interactions (DDIs) using machine learning has been studied extensively. However, most of the approaches have focused on text data or textual representation of the drug structures. We present the first work that uses multiple data sources such as drug structure images, drug structure string representation and relational representation of drug relationships as the input. To this effect, we exploit the recent advances in deep networks to integrate these varied sources of inputs in predicting DDIs. Our empirical evaluation against several state-of-the-art methods using standalone different data types for drugs clearly demonstrate the efficacy of combining heterogeneous data in predicting DDIs.
High-Throughput Approach to Modeling Healthcare Costs Using Electronic Healthcare Records
Nov 18, 2020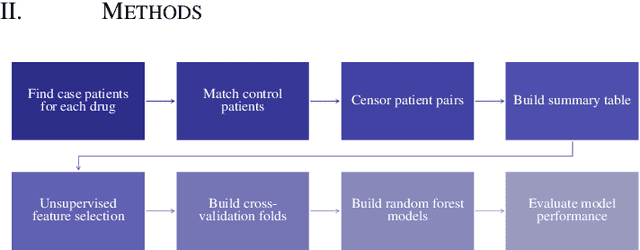
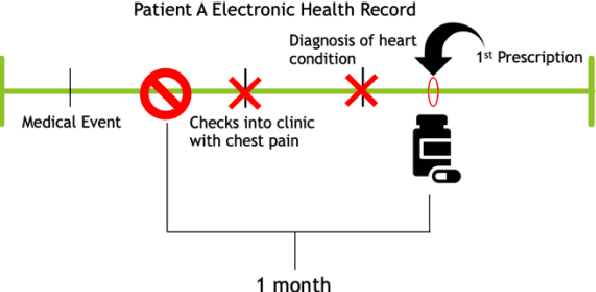
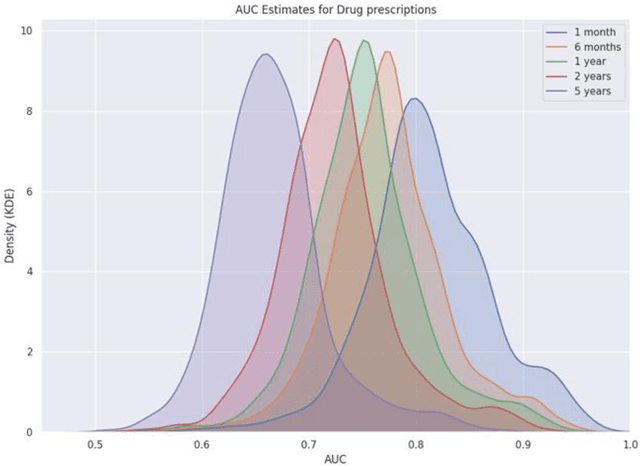
Abstract:Accurate estimation of healthcare costs is crucial for healthcare systems to plan and effectively negotiate with insurance companies regarding the coverage of patient-care costs. Greater accuracy in estimating healthcare costs would provide mutual benefit for both health systems and the insurers that support these systems by better aligning payment models with patient-care costs. This study presents the results of a generalizable machine learning approach to predicting medical events built from 40 years of data from >860,000 patients pertaining to >6,700 prescription medications, courtesy of Marshfield Clinic in Wisconsin. It was found that models built using this approach performed well when compared to similar studies predicting physician prescriptions of individual medications. In addition to providing a comprehensive predictive model for all drugs in a large healthcare system, the approach taken in this research benefits from potential applicability to a wide variety of other medical events.
Temporal Poisson Square Root Graphical Models
May 12, 2020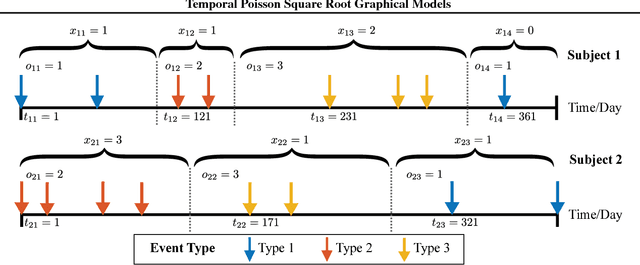
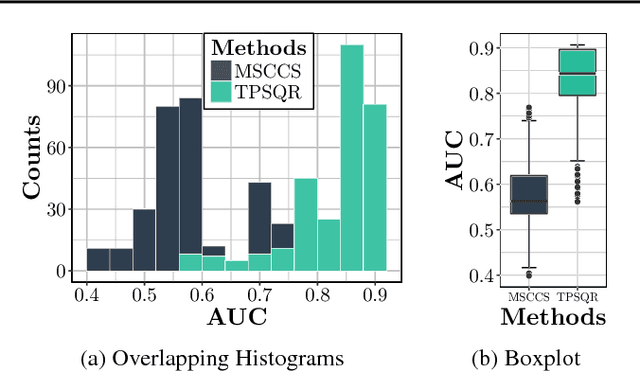
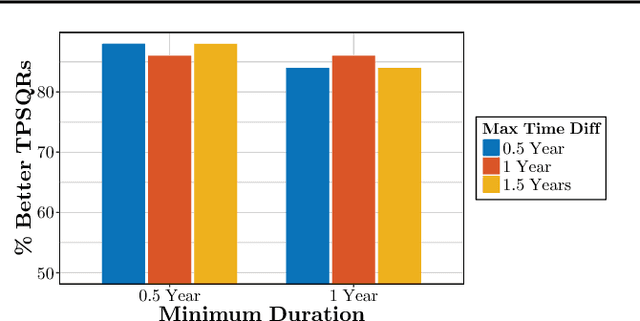
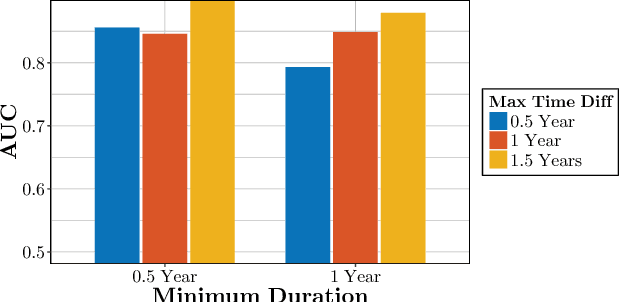
Abstract:We propose temporal Poisson square root graphical models (TPSQRs), a generalization of Poisson square root graphical models (PSQRs) specifically designed for modeling longitudinal event data. By estimating the temporal relationships for all possible pairs of event types, TPSQRs can offer a holistic perspective about whether the occurrences of any given event type could excite or inhibit any other type. A TPSQR is learned by estimating a collection of interrelated PSQRs that share the same template parameterization. These PSQRs are estimated jointly in a pseudo-likelihood fashion, where Poisson pseudo-likelihood is used to approximate the original more computationally-intensive pseudo-likelihood problem stemming from PSQRs. Theoretically, we demonstrate that under mild assumptions, the Poisson pseudo-likelihood approximation is sparsistent for recovering the underlying PSQR. Empirically, we learn TPSQRs from Marshfield Clinic electronic health records (EHRs) with millions of drug prescription and condition diagnosis events, for adverse drug reaction (ADR) detection. Experimental results demonstrate that the learned TPSQRs can recover ADR signals from the EHR effectively and efficiently.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge