Matthew M. Engelhard
Interval-Based AUC (iAUC): Extending ROC Analysis to Uncertainty-Aware Classification
Feb 04, 2026Abstract:In high-stakes risk prediction, quantifying uncertainty through interval-valued predictions is essential for reliable decision-making. However, standard evaluation tools like the receiver operating characteristic (ROC) curve and the area under the curve (AUC) are designed for point scores and fail to capture the impact of predictive uncertainty on ranking performance. We propose an uncertainty-aware ROC framework specifically for interval-valued predictions, introducing two new measures: $AUC_L$ and $AUC_U$. This framework enables an informative three-region decomposition of the ROC plane, partitioning pairwise rankings into correct, incorrect, and uncertain orderings. This approach naturally supports selective prediction by allowing models to abstain from ranking cases with overlapping intervals, thereby optimizing the trade-off between abstention rate and discriminative reliability. We prove that under valid class-conditional coverage, $AUC_L$ and $AUC_U$ provide formal lower and upper bounds on the theoretical optimal AUC ($AUC^*$), characterizing the physical limit of achievable discrimination. The proposed framework applies broadly to interval-valued prediction models, regardless of the interval construction method. Experiments on real-world benchmark datasets, using bootstrap-based intervals as one instantiation, validate the framework's correctness and demonstrate its practical utility for uncertainty-aware evaluation and decision-making.
FairPOT: Balancing AUC Performance and Fairness with Proportional Optimal Transport
Aug 05, 2025Abstract:Fairness metrics utilizing the area under the receiver operator characteristic curve (AUC) have gained increasing attention in high-stakes domains such as healthcare, finance, and criminal justice. In these domains, fairness is often evaluated over risk scores rather than binary outcomes, and a common challenge is that enforcing strict fairness can significantly degrade AUC performance. To address this challenge, we propose Fair Proportional Optimal Transport (FairPOT), a novel, model-agnostic post-processing framework that strategically aligns risk score distributions across different groups using optimal transport, but does so selectively by transforming a controllable proportion, i.e., the top-lambda quantile, of scores within the disadvantaged group. By varying lambda, our method allows for a tunable trade-off between reducing AUC disparities and maintaining overall AUC performance. Furthermore, we extend FairPOT to the partial AUC setting, enabling fairness interventions to concentrate on the highest-risk regions. Extensive experiments on synthetic, public, and clinical datasets show that FairPOT consistently outperforms existing post-processing techniques in both global and partial AUC scenarios, often achieving improved fairness with slight AUC degradation or even positive gains in utility. The computational efficiency and practical adaptability of FairPOT make it a promising solution for real-world deployment.
Infinite hierarchical contrastive clustering for personal digital envirotyping
May 21, 2025Abstract:Daily environments have profound influence on our health and behavior. Recent work has shown that digital envirotyping, where computer vision is applied to images of daily environments taken during ecological momentary assessment (EMA), can be used to identify meaningful relationships between environmental features and health outcomes of interest. To systematically study such effects on an individual level, it is helpful to group images into distinct environments encountered in an individual's daily life; these may then be analyzed, further grouped into related environments with similar features, and linked to health outcomes. Here we introduce infinite hierarchical contrastive clustering to address this challenge. Building on the established contrastive clustering framework, our method a) allows an arbitrary number of clusters without requiring the full Dirichlet Process machinery by placing a stick-breaking prior on predicted cluster probabilities; and b) encourages distinct environments to form well-defined sub-clusters within each cluster of related environments by incorporating a participant-specific prediction loss. Our experiments show that our model effectively identifies distinct personal environments and groups these environments into meaningful environment types. We then illustrate how the resulting clusters can be linked to various health outcomes, highlighting the potential of our approach to advance the envirotyping paradigm.
MALADE: Orchestration of LLM-powered Agents with Retrieval Augmented Generation for Pharmacovigilance
Aug 03, 2024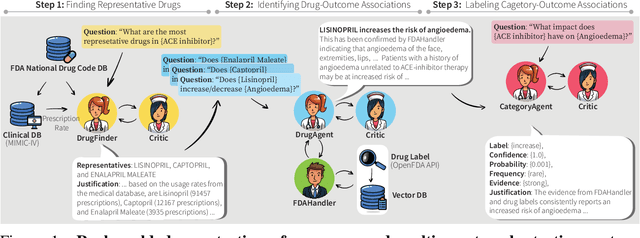
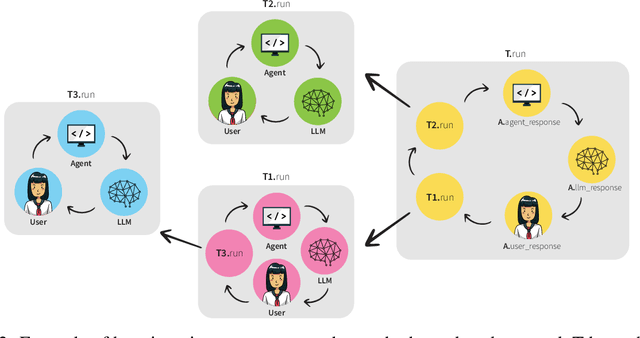
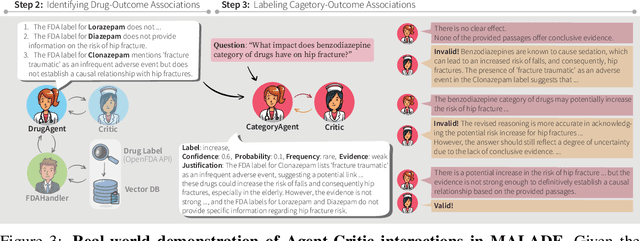
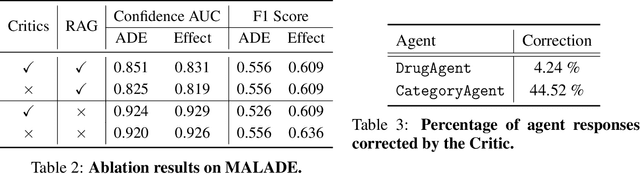
Abstract:In the era of Large Language Models (LLMs), given their remarkable text understanding and generation abilities, there is an unprecedented opportunity to develop new, LLM-based methods for trustworthy medical knowledge synthesis, extraction and summarization. This paper focuses on the problem of Pharmacovigilance (PhV), where the significance and challenges lie in identifying Adverse Drug Events (ADEs) from diverse text sources, such as medical literature, clinical notes, and drug labels. Unfortunately, this task is hindered by factors including variations in the terminologies of drugs and outcomes, and ADE descriptions often being buried in large amounts of narrative text. We present MALADE, the first effective collaborative multi-agent system powered by LLM with Retrieval Augmented Generation for ADE extraction from drug label data. This technique involves augmenting a query to an LLM with relevant information extracted from text resources, and instructing the LLM to compose a response consistent with the augmented data. MALADE is a general LLM-agnostic architecture, and its unique capabilities are: (1) leveraging a variety of external sources, such as medical literature, drug labels, and FDA tools (e.g., OpenFDA drug information API), (2) extracting drug-outcome association in a structured format along with the strength of the association, and (3) providing explanations for established associations. Instantiated with GPT-4 Turbo or GPT-4o, and FDA drug label data, MALADE demonstrates its efficacy with an Area Under ROC Curve of 0.90 against the OMOP Ground Truth table of ADEs. Our implementation leverages the Langroid multi-agent LLM framework and can be found at https://github.com/jihyechoi77/malade.
Improving Event Time Prediction by Learning to Partition the Event Time Space
Oct 24, 2023



Abstract:Recently developed survival analysis methods improve upon existing approaches by predicting the probability of event occurrence in each of a number pre-specified (discrete) time intervals. By avoiding placing strong parametric assumptions on the event density, this approach tends to improve prediction performance, particularly when data are plentiful. However, in clinical settings with limited available data, it is often preferable to judiciously partition the event time space into a limited number of intervals well suited to the prediction task at hand. In this work, we develop a method to learn from data a set of cut points defining such a partition. We show that in two simulated datasets, we are able to recover intervals that match the underlying generative model. We then demonstrate improved prediction performance on three real-world observational datasets, including a large, newly harmonized stroke risk prediction dataset. Finally, we argue that our approach facilitates clinical decision-making by suggesting time intervals that are most appropriate for each task, in the sense that they facilitate more accurate risk prediction.
On Neural Networks as Infinite Tree-Structured Probabilistic Graphical Models
May 27, 2023Abstract:Deep neural networks (DNNs) lack the precise semantics and definitive probabilistic interpretation of probabilistic graphical models (PGMs). In this paper, we propose an innovative solution by constructing infinite tree-structured PGMs that correspond exactly to neural networks. Our research reveals that DNNs, during forward propagation, indeed perform approximations of PGM inference that are precise in this alternative PGM structure. Not only does our research complement existing studies that describe neural networks as kernel machines or infinite-sized Gaussian processes, it also elucidates a more direct approximation that DNNs make to exact inference in PGMs. Potential benefits include improved pedagogy and interpretation of DNNs, and algorithms that can merge the strengths of PGMs and DNNs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge