Bradford J. Wood
OpenPros: A Large-Scale Dataset for Limited View Prostate Ultrasound Computed Tomography
May 18, 2025Abstract:Prostate cancer is one of the most common and lethal cancers among men, making its early detection critically important. Although ultrasound imaging offers greater accessibility and cost-effectiveness compared to MRI, traditional transrectal ultrasound methods suffer from low sensitivity, especially in detecting anteriorly located tumors. Ultrasound computed tomography provides quantitative tissue characterization, but its clinical implementation faces significant challenges, particularly under anatomically constrained limited-angle acquisition conditions specific to prostate imaging. To address these unmet needs, we introduce OpenPros, the first large-scale benchmark dataset explicitly developed for limited-view prostate USCT. Our dataset includes over 280,000 paired samples of realistic 2D speed-of-sound (SOS) phantoms and corresponding ultrasound full-waveform data, generated from anatomically accurate 3D digital prostate models derived from real clinical MRI/CT scans and ex vivo ultrasound measurements, annotated by medical experts. Simulations are conducted under clinically realistic configurations using advanced finite-difference time-domain and Runge-Kutta acoustic wave solvers, both provided as open-source components. Through comprehensive baseline experiments, we demonstrate that state-of-the-art deep learning methods surpass traditional physics-based approaches in both inference efficiency and reconstruction accuracy. Nevertheless, current deep learning models still fall short of delivering clinically acceptable high-resolution images with sufficient accuracy. By publicly releasing OpenPros, we aim to encourage the development of advanced machine learning algorithms capable of bridging this performance gap and producing clinically usable, high-resolution, and highly accurate prostate ultrasound images. The dataset is publicly accessible at https://open-pros.github.io/.
C-DARL: Contrastive diffusion adversarial representation learning for label-free blood vessel segmentation
Jul 31, 2023Abstract:Blood vessel segmentation in medical imaging is one of the essential steps for vascular disease diagnosis and interventional planning in a broad spectrum of clinical scenarios in image-based medicine and interventional medicine. Unfortunately, manual annotation of the vessel masks is challenging and resource-intensive due to subtle branches and complex structures. To overcome this issue, this paper presents a self-supervised vessel segmentation method, dubbed the contrastive diffusion adversarial representation learning (C-DARL) model. Our model is composed of a diffusion module and a generation module that learns the distribution of multi-domain blood vessel data by generating synthetic vessel images from diffusion latent. Moreover, we employ contrastive learning through a mask-based contrastive loss so that the model can learn more realistic vessel representations. To validate the efficacy, C-DARL is trained using various vessel datasets, including coronary angiograms, abdominal digital subtraction angiograms, and retinal imaging. Experimental results confirm that our model achieves performance improvement over baseline methods with noise robustness, suggesting the effectiveness of C-DARL for vessel segmentation.
Distance Map Supervised Landmark Localization for MR-TRUS Registration
Oct 11, 2022


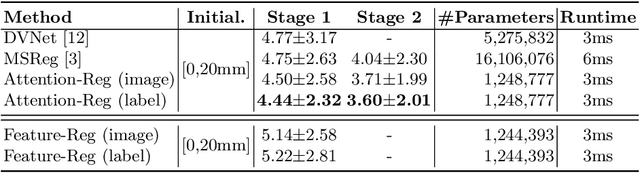
Abstract:In this work, we propose to explicitly use the landmarks of prostate to guide the MR-TRUS image registration. We first train a deep neural network to automatically localize a set of meaningful landmarks, and then directly generate the affine registration matrix from the location of these landmarks. For landmark localization, instead of directly training a network to predict the landmark coordinates, we propose to regress a full-resolution distance map of the landmark, which is demonstrated effective in avoiding statistical bias to unsatisfactory performance and thus improving performance. We then use the predicted landmarks to generate the affine transformation matrix, which outperforms the clinicians' manual rigid registration by a significant margin in terms of TRE.
End-to-end Ultrasound Frame to Volume Registration
Jul 14, 2021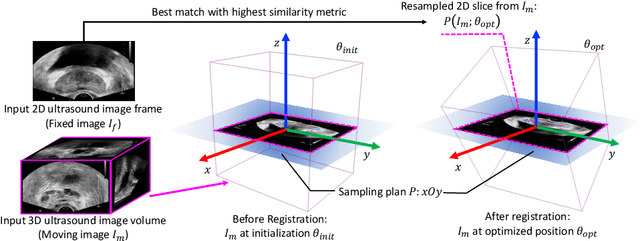
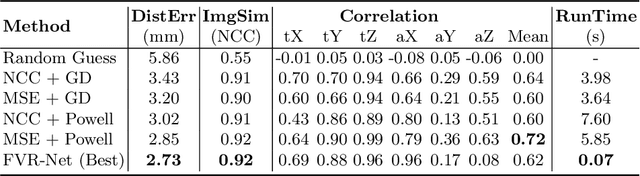
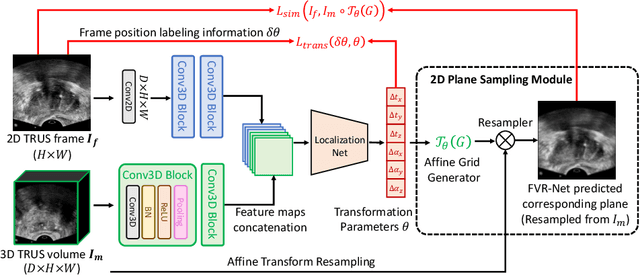
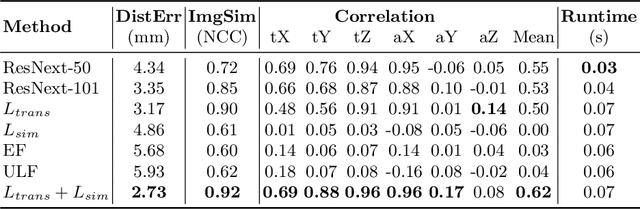
Abstract:Fusing intra-operative 2D transrectal ultrasound (TRUS) image with pre-operative 3D magnetic resonance (MR) volume to guide prostate biopsy can significantly increase the yield. However, such a multimodal 2D/3D registration problem is a very challenging task. In this paper, we propose an end-to-end frame-to-volume registration network (FVR-Net), which can efficiently bridge the previous research gaps by aligning a 2D TRUS frame with a 3D TRUS volume without requiring hardware tracking. The proposed FVR-Net utilizes a dual-branch feature extraction module to extract the information from TRUS frame and volume to estimate transformation parameters. We also introduce a differentiable 2D slice sampling module which allows gradients backpropagating from an unsupervised image similarity loss for content correspondence learning. Our model shows superior efficiency for real-time interventional guidance with highly competitive registration accuracy.
Cross-modal Attention for MRI and Ultrasound Volume Registration
Jul 12, 2021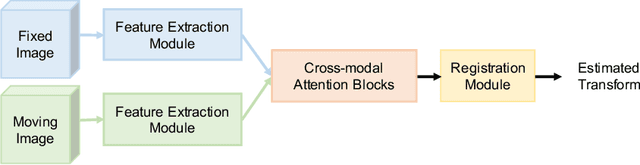


Abstract:Prostate cancer biopsy benefits from accurate fusion of transrectal ultrasound (TRUS) and magnetic resonance (MR) images. In the past few years, convolutional neural networks (CNNs) have been proved powerful in extracting image features crucial for image registration. However, challenging applications and recent advances in computer vision suggest that CNNs are quite limited in its ability to understand spatial correspondence between features, a task in which the self-attention mechanism excels. This paper aims to develop a self-attention mechanism specifically for cross-modal image registration. Our proposed cross-modal attention block effectively maps each of the features in one volume to all features in the corresponding volume. Our experimental results demonstrate that a CNN network designed with the cross-modal attention block embedded outperforms an advanced CNN network 10 times of its size. We also incorporated visualization techniques to improve the interpretability of our network. The source code of our work is available at https://github.com/DIAL-RPI/Attention-Reg .
Federated Semi-Supervised Learning for COVID Region Segmentation in Chest CT using Multi-National Data from China, Italy, Japan
Nov 23, 2020



Abstract:The recent outbreak of COVID-19 has led to urgent needs for reliable diagnosis and management of SARS-CoV-2 infection. As a complimentary tool, chest CT has been shown to be able to reveal visual patterns characteristic for COVID-19, which has definite value at several stages during the disease course. To facilitate CT analysis, recent efforts have focused on computer-aided characterization and diagnosis, which has shown promising results. However, domain shift of data across clinical data centers poses a serious challenge when deploying learning-based models. In this work, we attempt to find a solution for this challenge via federated and semi-supervised learning. A multi-national database consisting of 1704 scans from three countries is adopted to study the performance gap, when training a model with one dataset and applying it to another. Expert radiologists manually delineated 945 scans for COVID-19 findings. In handling the variability in both the data and annotations, a novel federated semi-supervised learning technique is proposed to fully utilize all available data (with or without annotations). Federated learning avoids the need for sensitive data-sharing, which makes it favorable for institutions and nations with strict regulatory policy on data privacy. Moreover, semi-supervision potentially reduces the annotation burden under a distributed setting. The proposed framework is shown to be effective compared to fully supervised scenarios with conventional data sharing instead of model weight sharing.
Transducer Adaptive Ultrasound Volume Reconstruction
Nov 17, 2020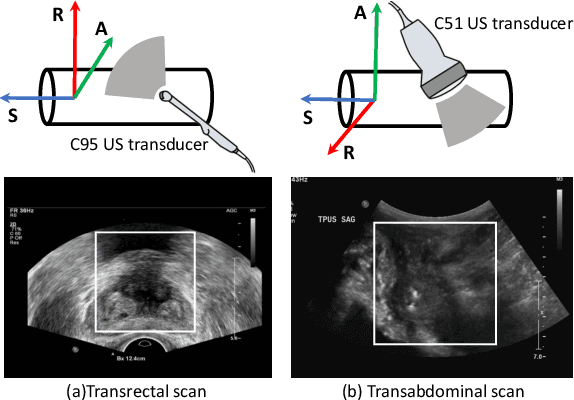



Abstract:Reconstructed 3D ultrasound volume provides more context information compared to a sequence of 2D scanning frames, which is desirable for various clinical applications such as ultrasound-guided prostate biopsy. Nevertheless, 3D volume reconstruction from freehand 2D scans is a very challenging problem, especially without the use of external tracking devices. Recent deep learning based methods demonstrate the potential of directly estimating inter-frame motion between consecutive ultrasound frames. However, such algorithms are specific to particular transducers and scanning trajectories associated with the training data, which may not be generalized to other image acquisition settings. In this paper, we tackle the data acquisition difference as a domain shift problem and propose a novel domain adaptation strategy to adapt deep learning algorithms to data acquired with different transducers. Specifically, feature extractors that generate transducer-invariant features from different datasets are trained by minimizing the discrepancy between deep features of paired samples in a latent space. Our results show that the proposed domain adaptation method can successfully align different feature distributions while preserving the transducer-specific information for universal freehand ultrasound volume reconstruction.
Unified Multi-scale Feature Abstraction for Medical Image Segmentation
Oct 24, 2019



Abstract:Automatic medical image segmentation, an essential component of medical image analysis, plays an importantrole in computer-aided diagnosis. For example, locating and segmenting the liver can be very helpful in livercancer diagnosis and treatment. The state-of-the-art models in medical image segmentation are variants ofthe encoder-decoder architecture such as fully convolutional network (FCN) and U-Net.1A major focus ofthe FCN based segmentation methods has been on network structure engineering by incorporating the latestCNN structures such as ResNet2and DenseNet.3In addition to exploring new network structures for efficientlyabstracting high level features, incorporating structures for multi-scale image feature extraction in FCN hashelped to improve performance in segmentation tasks. In this paper, we design a new multi-scale networkarchitecture, which takes multi-scale inputs with dedicated convolutional paths to efficiently combine featuresfrom different scales to better utilize the hierarchical information.
Gaze2Segment: A Pilot Study for Integrating Eye-Tracking Technology into Medical Image Segmentation
Aug 10, 2016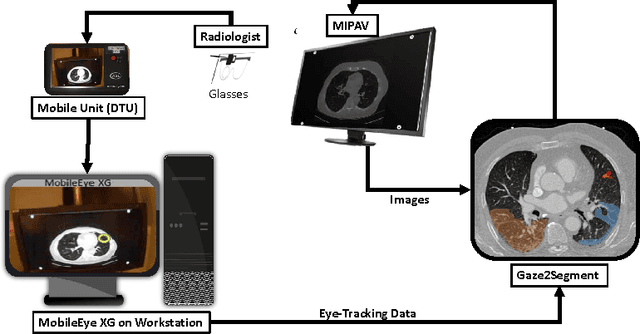
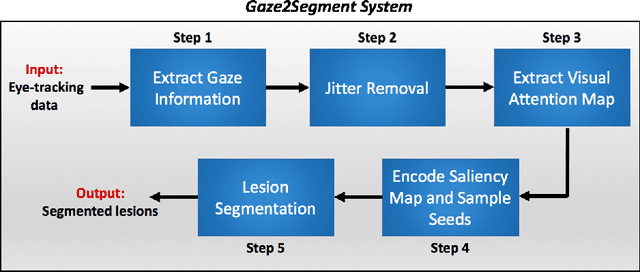
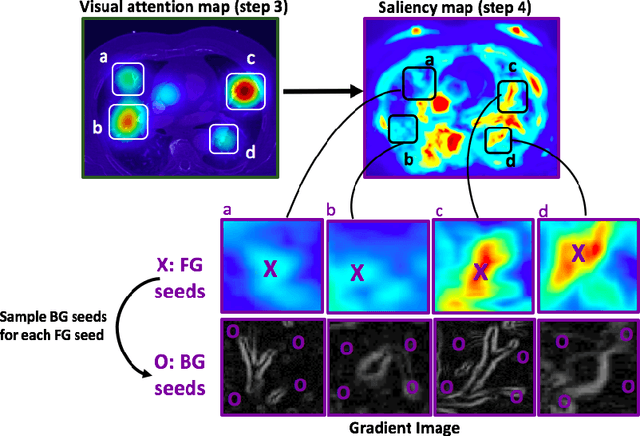
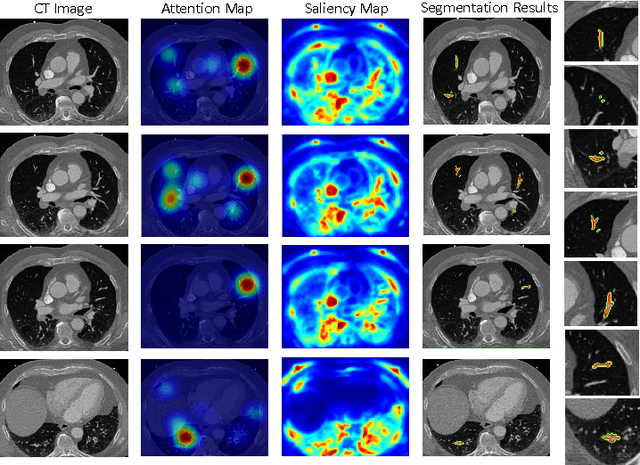
Abstract:This study introduced a novel system, called Gaze2Segment, integrating biological and computer vision techniques to support radiologists' reading experience with an automatic image segmentation task. During diagnostic assessment of lung CT scans, the radiologists' gaze information were used to create a visual attention map. This map was then combined with a computer-derived saliency map, extracted from the gray-scale CT images. The visual attention map was used as an input for indicating roughly the location of a object of interest. With computer-derived saliency information, on the other hand, we aimed at finding foreground and background cues for the object of interest. At the final step, these cues were used to initiate a seed-based delineation process. Segmentation accuracy of the proposed Gaze2Segment was found to be 86% with dice similarity coefficient and 1.45 mm with Hausdorff distance. To the best of our knowledge, Gaze2Segment is the first true integration of eye-tracking technology into a medical image segmentation task without the need for any further user-interaction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge