Hengtao Guo
Independent Researcher, Seattle, WA, US
Retinal Vessel Segmentation via Neuron Programming
Nov 17, 2024
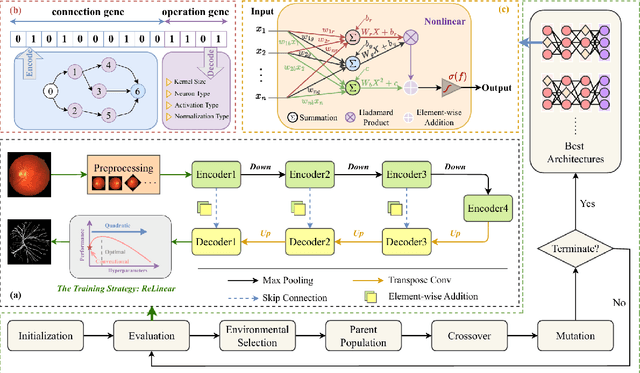
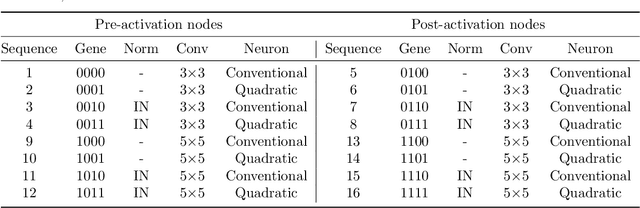
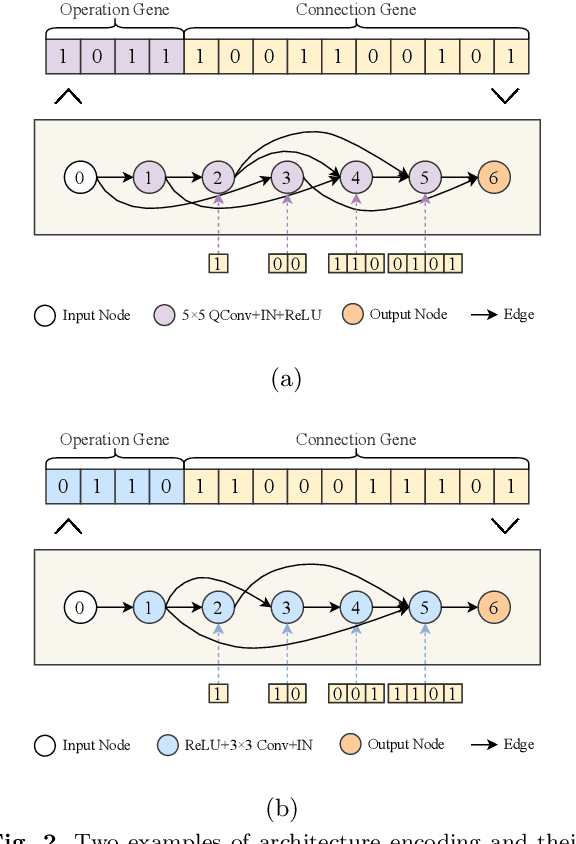
Abstract:The accurate segmentation of retinal blood vessels plays a crucial role in the early diagnosis and treatment of various ophthalmic diseases. Designing a network model for this task requires meticulous tuning and extensive experimentation to handle the tiny and intertwined morphology of retinal blood vessels. To tackle this challenge, Neural Architecture Search (NAS) methods are developed to fully explore the space of potential network architectures and go after the most powerful one. Inspired by neuronal diversity which is the biological foundation of all kinds of intelligent behaviors in our brain, this paper introduces a novel and foundational approach to neural network design, termed ``neuron programming'', to automatically search neuronal types into a network to enhance a network's representation ability at the neuronal level, which is complementary to architecture-level enhancement done by NAS. Additionally, to mitigate the time and computational intensity of neuron programming, we develop a hypernetwork that leverages the search-derived architectural information to predict optimal neuronal configurations. Comprehensive experiments validate that neuron programming can achieve competitive performance in retinal blood segmentation, demonstrating the strong potential of neuronal diversity in medical image analysis.
Don't Fear Peculiar Activation Functions: EUAF and Beyond
Jul 12, 2024



Abstract:In this paper, we propose a new super-expressive activation function called the Parametric Elementary Universal Activation Function (PEUAF). We demonstrate the effectiveness of PEUAF through systematic and comprehensive experiments on various industrial and image datasets, including CIFAR10, Tiny-ImageNet, and ImageNet. Moreover, we significantly generalize the family of super-expressive activation functions, whose existence has been demonstrated in several recent works by showing that any continuous function can be approximated to any desired accuracy by a fixed-size network with a specific super-expressive activation function. Specifically, our work addresses two major bottlenecks in impeding the development of super-expressive activation functions: the limited identification of super-expressive functions, which raises doubts about their broad applicability, and their often peculiar forms, which lead to skepticism regarding their scalability and practicality in real-world applications.
End-to-end Ultrasound Frame to Volume Registration
Jul 14, 2021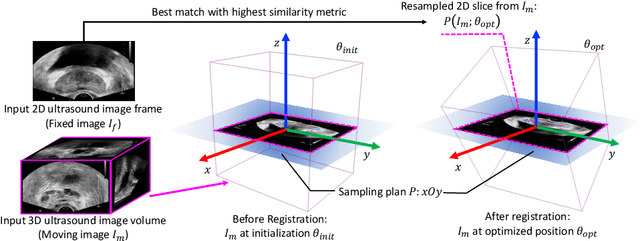
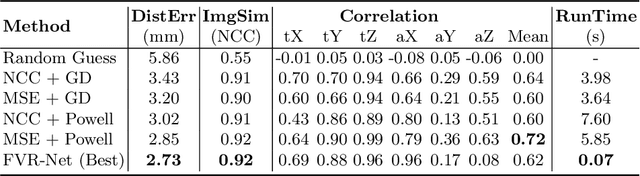
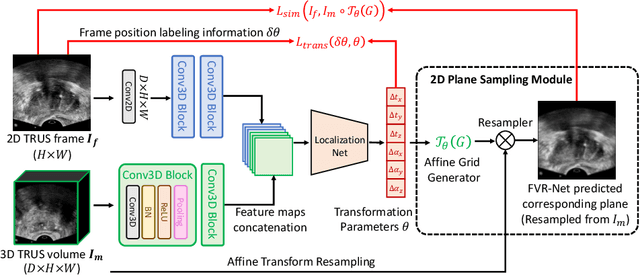
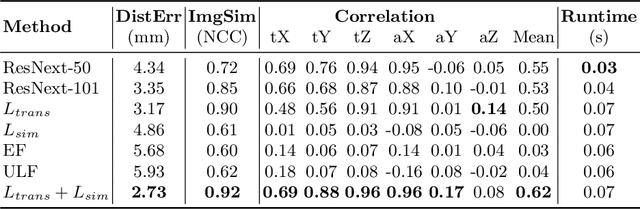
Abstract:Fusing intra-operative 2D transrectal ultrasound (TRUS) image with pre-operative 3D magnetic resonance (MR) volume to guide prostate biopsy can significantly increase the yield. However, such a multimodal 2D/3D registration problem is a very challenging task. In this paper, we propose an end-to-end frame-to-volume registration network (FVR-Net), which can efficiently bridge the previous research gaps by aligning a 2D TRUS frame with a 3D TRUS volume without requiring hardware tracking. The proposed FVR-Net utilizes a dual-branch feature extraction module to extract the information from TRUS frame and volume to estimate transformation parameters. We also introduce a differentiable 2D slice sampling module which allows gradients backpropagating from an unsupervised image similarity loss for content correspondence learning. Our model shows superior efficiency for real-time interventional guidance with highly competitive registration accuracy.
Cross-modal Attention for MRI and Ultrasound Volume Registration
Jul 12, 2021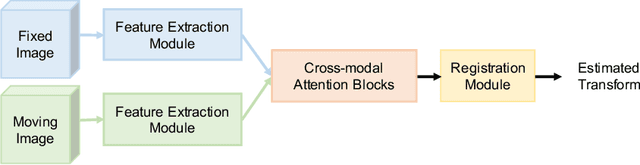


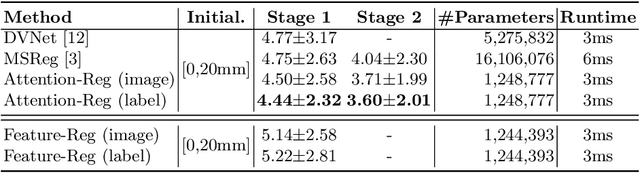
Abstract:Prostate cancer biopsy benefits from accurate fusion of transrectal ultrasound (TRUS) and magnetic resonance (MR) images. In the past few years, convolutional neural networks (CNNs) have been proved powerful in extracting image features crucial for image registration. However, challenging applications and recent advances in computer vision suggest that CNNs are quite limited in its ability to understand spatial correspondence between features, a task in which the self-attention mechanism excels. This paper aims to develop a self-attention mechanism specifically for cross-modal image registration. Our proposed cross-modal attention block effectively maps each of the features in one volume to all features in the corresponding volume. Our experimental results demonstrate that a CNN network designed with the cross-modal attention block embedded outperforms an advanced CNN network 10 times of its size. We also incorporated visualization techniques to improve the interpretability of our network. The source code of our work is available at https://github.com/DIAL-RPI/Attention-Reg .
Transducer Adaptive Ultrasound Volume Reconstruction
Nov 17, 2020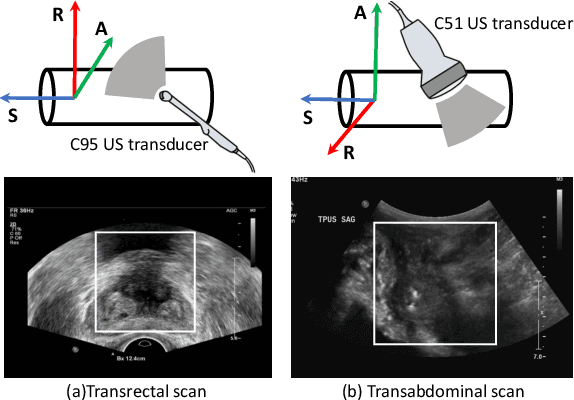



Abstract:Reconstructed 3D ultrasound volume provides more context information compared to a sequence of 2D scanning frames, which is desirable for various clinical applications such as ultrasound-guided prostate biopsy. Nevertheless, 3D volume reconstruction from freehand 2D scans is a very challenging problem, especially without the use of external tracking devices. Recent deep learning based methods demonstrate the potential of directly estimating inter-frame motion between consecutive ultrasound frames. However, such algorithms are specific to particular transducers and scanning trajectories associated with the training data, which may not be generalized to other image acquisition settings. In this paper, we tackle the data acquisition difference as a domain shift problem and propose a novel domain adaptation strategy to adapt deep learning algorithms to data acquired with different transducers. Specifically, feature extractors that generate transducer-invariant features from different datasets are trained by minimizing the discrepancy between deep features of paired samples in a latent space. Our results show that the proposed domain adaptation method can successfully align different feature distributions while preserving the transducer-specific information for universal freehand ultrasound volume reconstruction.
Deep Learning Predicts Cardiovascular Disease Risks from Lung Cancer Screening Low Dose Computed Tomography
Aug 16, 2020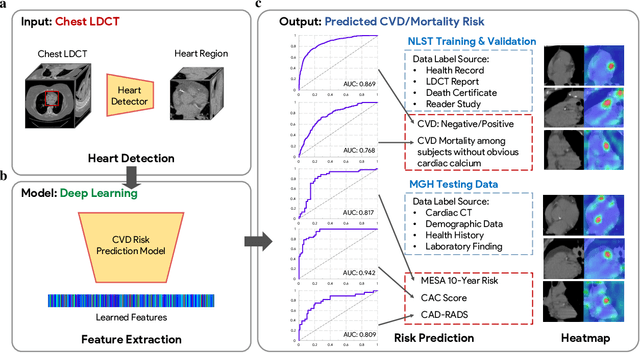

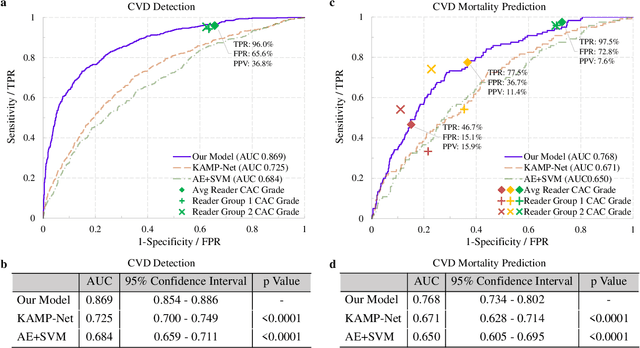

Abstract:The high risk population of cardiovascular disease (CVD) is simultaneously at high risk of lung cancer. Given the dominance of low dose computed tomography (LDCT) for lung cancer screening, the feasibility of extracting information on CVD from the same LDCT scan would add major value to patients at no additional radiation dose. However, with strong noise in LDCT images and without electrocardiogram (ECG) gating, CVD risk analysis from LDCT is highly challenging. Here we present an innovative deep learning model to address this challenge. Our deep model was trained with 30,286 LDCT volumes and achieved the state-of-the-art performance (area under the curve (AUC) of 0.869) on 2,085 National Lung Cancer Screening Trial (NLST) subjects, and effectively identified patients with high CVD mortality risks (AUC of 0.768). Our deep model was further calibrated against the clinical gold standard CVD risk scores from ECG-gated dedicated cardiac CT, including coronary artery calcification (CAC) score, CAD-RADS score and MESA 10-year CHD risk score from an independent dataset of 106 subjects. In this validation study, our model achieved AUC of 0.942, 0.809 and 0.817 for CAC, CAD-RADS and MESA scores, respectively. Our deep learning model has the potential to convert LDCT for lung cancer screening into dual-screening quantitative tool for CVD risk estimation.
Sensorless Freehand 3D Ultrasound Reconstruction via Deep Contextual Learning
Jun 13, 2020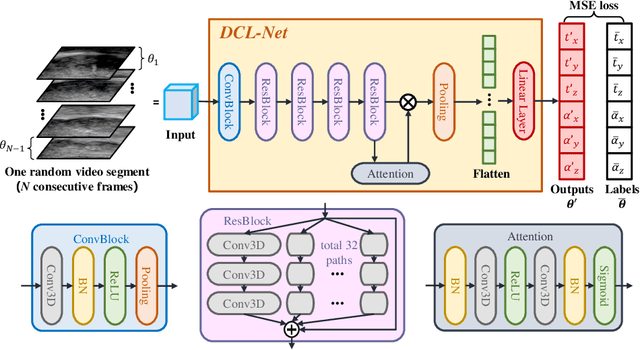
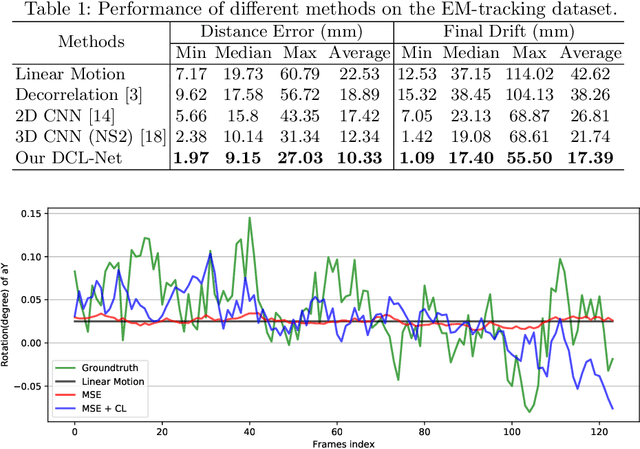

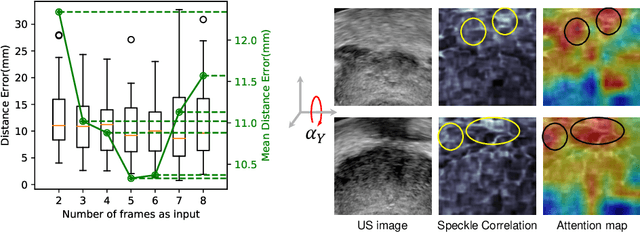
Abstract:Transrectal ultrasound (US) is the most commonly used imaging modality to guide prostate biopsy and its 3D volume provides even richer context information. Current methods for 3D volume reconstruction from freehand US scans require external tracking devices to provide spatial position for every frame. In this paper, we propose a deep contextual learning network (DCL-Net), which can efficiently exploit the image feature relationship between US frames and reconstruct 3D US volumes without any tracking device. The proposed DCL-Net utilizes 3D convolutions over a US video segment for feature extraction. An embedded self-attention module makes the network focus on the speckle-rich areas for better spatial movement prediction. We also propose a novel case-wise correlation loss to stabilize the training process for improved accuracy. Highly promising results have been obtained by using the developed method. The experiments with ablation studies demonstrate superior performance of the proposed method by comparing against other state-of-the-art methods. Source code of this work is publicly available at https://github.com/DIAL-RPI/FreehandUSRecon.
Knowledge-based Analysis for Mortality Prediction from CT Images
Feb 20, 2019
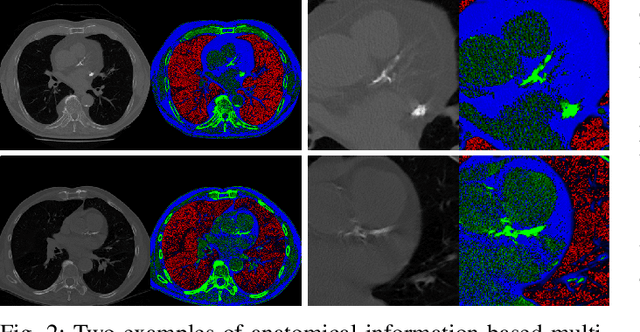


Abstract:Recent studies have highlighted the high correlation between cardiovascular diseases (CVD) and lung cancer, and both are associated with significant morbidity and mortality. Low-Dose CT (LCDT) scans have led to significant improvements in the accuracy of lung cancer diagnosis and thus the reduction of cancer deaths. However, the high correlation between lung cancer and CVD has not been well explored for mortality prediction. This paper introduces a knowledge-based analytical method using deep convolutional neural network (CNN) for all-cause mortality prediction. The underlying approach combines structural image features extracted from CNNs, based on LDCT volume in different scale, and clinical knowledge obtained from quantitative measurements, to comprehensively predict the mortality risk of lung cancer screening subjects. The introduced method is referred to here as the Knowledge-based Analysis of Mortality Prediction Network, or KAMP-Net. It constitutes a collaborative framework that utilizes both imaging features and anatomical information, instead of completely relying on automatic feature extraction. Our work demonstrates the feasibility of incorporating quantitative clinical measurements to assist CNNs in all-cause mortality prediction from chest LDCT images. The results of this study confirm that radiologist defined features are an important complement to CNNs to achieve a more comprehensive feature extraction. Thus, the proposed KAMP-Net has shown to achieve a superior performance when compared to other methods.
Hybrid deep neural networks for all-cause Mortality Prediction from LDCT Images
Oct 19, 2018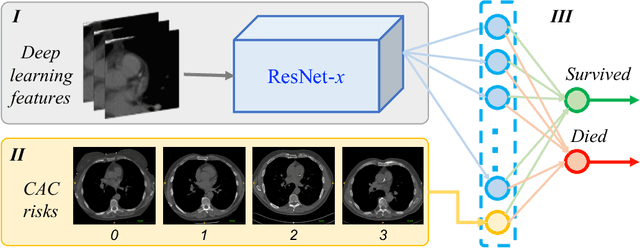
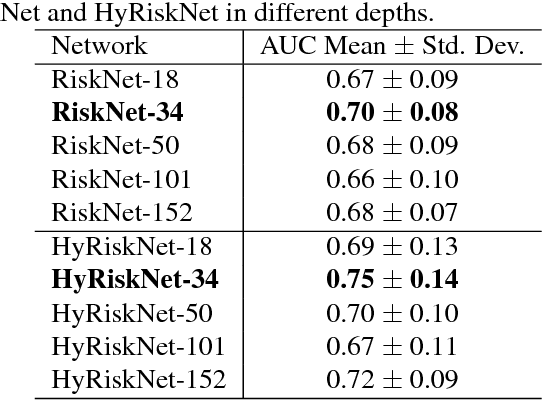
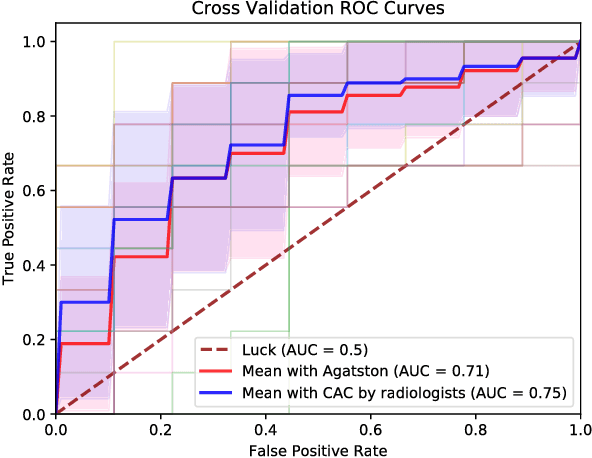
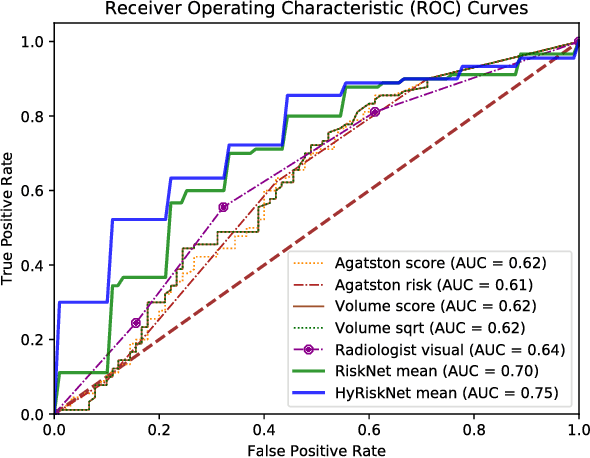
Abstract:Known for its high morbidity and mortality rates, lung cancer poses a significant threat to human health and well-being. However, the same population is also at high risk for other deadly diseases, such as cardiovascular disease. Since Low-Dose CT (LDCT) has been shown to significantly improve the lung cancer diagnosis accuracy, it will be very useful for clinical practice to predict the all-cause mortality for lung cancer patients to take corresponding actions. In this paper, we propose a deep learning based method, which takes both chest LDCT image patches and coronary artery calcification risk scores as input, for direct prediction of mortality risk of lung cancer subjects. The proposed method is called Hybrid Risk Network (HyRiskNet) for mortality risk prediction, which is an end-to-end framework utilizing hybrid imaging features, instead of completely relying on automatic feature extraction. Our work demonstrates the feasibility of using deep learning techniques for all-cause lung cancer mortality prediction from chest LDCT images. The experimental results show that the proposed HyRiskNet can achieve superior performance compared with the neural networks with only image input and with other traditional semi-automatic scoring methods. The study also indicates that radiologist defined features can well complement convolutional neural networks for more comprehensive feature extraction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge