Zimeng Li
GSR: Learning Structured Reasoning for Embodied Manipulation
Feb 02, 2026Abstract:Despite rapid progress, embodied agents still struggle with long-horizon manipulation that requires maintaining spatial consistency, causal dependencies, and goal constraints. A key limitation of existing approaches is that task reasoning is implicitly embedded in high-dimensional latent representations, making it challenging to separate task structure from perceptual variability. We introduce Grounded Scene-graph Reasoning (GSR), a structured reasoning paradigm that explicitly models world-state evolution as transitions over semantically grounded scene graphs. By reasoning step-wise over object states and spatial relations, rather than directly mapping perception to actions, GSR enables explicit reasoning about action preconditions, consequences, and goal satisfaction in a physically grounded space. To support learning such reasoning, we construct Manip-Cognition-1.6M, a large-scale dataset that jointly supervises world understanding, action planning, and goal interpretation. Extensive evaluations across RLBench, LIBERO, GSR-benchmark, and real-world robotic tasks show that GSR significantly improves zero-shot generalization and long-horizon task completion over prompting-based baselines. These results highlight explicit world-state representations as a key inductive bias for scalable embodied reasoning.
ProMist-5K: A Comprehensive Dataset for Digital Emulation of Cinematic Pro-Mist Filter Effects
Jan 27, 2026Abstract:Pro-Mist filters are widely used in cinematography for their ability to create soft halation, lower contrast, and produce a distinctive, atmospheric style. These effects are difficult to reproduce digitally due to the complex behavior of light diffusion. We present ProMist-5K, a dataset designed to support cinematic style emulation. It is built using a physically inspired pipeline in a scene-referred linear space and includes 20,000 high-resolution image pairs across four configurations, covering two filter densities (1/2 and 1/8) and two focal lengths (20mm and 50mm). Unlike general style datasets, ProMist-5K focuses on realistic glow and highlight diffusion effects. Multiple blur layers and carefully tuned weighting are used to model the varying intensity and spread of optical diffusion. The dataset provides a consistent and controllable target domain that supports various image translation models and learning paradigms. Experiments show that the dataset works well across different training settings and helps capture both subtle and strong cinematic appearances. ProMist-5K offers a practical and physically grounded resource for film-inspired image transformation, bridging the gap between digital flexibility and traditional lens aesthetics. The dataset is available at https://www.kaggle.com/datasets/yingtielei/promist5k.
SFormer: SNR-guided Transformer for Underwater Image Enhancement from the Frequency Domain
Aug 26, 2025



Abstract:Recent learning-based underwater image enhancement (UIE) methods have advanced by incorporating physical priors into deep neural networks, particularly using the signal-to-noise ratio (SNR) prior to reduce wavelength-dependent attenuation. However, spatial domain SNR priors have two limitations: (i) they cannot effectively separate cross-channel interference, and (ii) they provide limited help in amplifying informative structures while suppressing noise. To overcome these, we propose using the SNR prior in the frequency domain, decomposing features into amplitude and phase spectra for better channel modulation. We introduce the Fourier Attention SNR-prior Transformer (FAST), combining spectral interactions with SNR cues to highlight key spectral components. Additionally, the Frequency Adaptive Transformer (FAT) bottleneck merges low- and high-frequency branches using a gated attention mechanism to enhance perceptual quality. Embedded in a unified U-shaped architecture, these modules integrate a conventional RGB stream with an SNR-guided branch, forming SFormer. Trained on 4,800 paired images from UIEB, EUVP, and LSUI, SFormer surpasses recent methods with a 3.1 dB gain in PSNR and 0.08 in SSIM, successfully restoring colors, textures, and contrast in underwater scenes.
CMAMRNet: A Contextual Mask-Aware Network Enhancing Mural Restoration Through Comprehensive Mask Guidance
Aug 10, 2025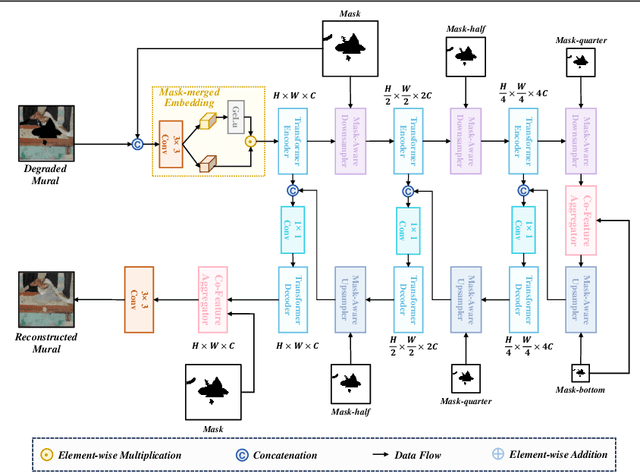
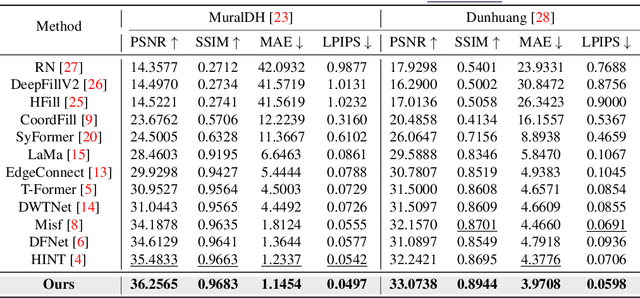
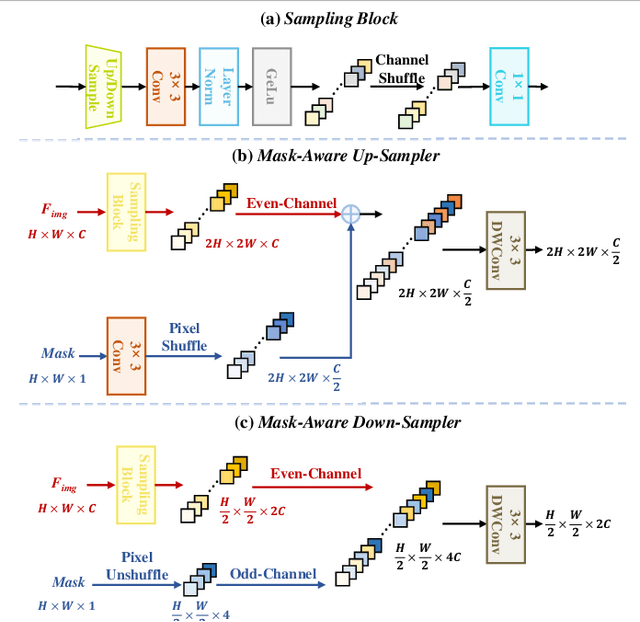
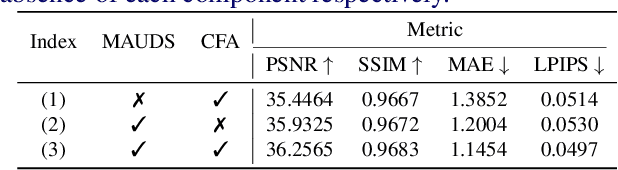
Abstract:Murals, as invaluable cultural artifacts, face continuous deterioration from environmental factors and human activities. Digital restoration of murals faces unique challenges due to their complex degradation patterns and the critical need to preserve artistic authenticity. Existing learning-based methods struggle with maintaining consistent mask guidance throughout their networks, leading to insufficient focus on damaged regions and compromised restoration quality. We propose CMAMRNet, a Contextual Mask-Aware Mural Restoration Network that addresses these limitations through comprehensive mask guidance and multi-scale feature extraction. Our framework introduces two key components: (1) the Mask-Aware Up/Down-Sampler (MAUDS), which ensures consistent mask sensitivity across resolution scales through dedicated channel-wise feature selection and mask-guided feature fusion; and (2) the Co-Feature Aggregator (CFA), operating at both the highest and lowest resolutions to extract complementary features for capturing fine textures and global structures in degraded regions. Experimental results on benchmark datasets demonstrate that CMAMRNet outperforms state-of-the-art methods, effectively preserving both structural integrity and artistic details in restored murals. The code is available at~\href{https://github.com/CXH-Research/CMAMRNet}{https://github.com/CXH-Research/CMAMRNet}.
Underwater Image Restoration via Polymorphic Large Kernel CNNs
Dec 24, 2024



Abstract:Underwater Image Restoration (UIR) remains a challenging task in computer vision due to the complex degradation of images in underwater environments. While recent approaches have leveraged various deep learning techniques, including Transformers and complex, parameter-heavy models to achieve significant improvements in restoration effects, we demonstrate that pure CNN architectures with lightweight parameters can achieve comparable results. In this paper, we introduce UIR-PolyKernel, a novel method for underwater image restoration that leverages Polymorphic Large Kernel CNNs. Our approach uniquely combines large kernel convolutions of diverse sizes and shapes to effectively capture long-range dependencies within underwater imagery. Additionally, we introduce a Hybrid Domain Attention module that integrates frequency and spatial domain attention mechanisms to enhance feature importance. By leveraging the frequency domain, we can capture hidden features that may not be perceptible to humans but are crucial for identifying patterns in both underwater and on-air images. This approach enhances the generalization and robustness of our UIR model. Extensive experiments on benchmark datasets demonstrate that UIR-PolyKernel achieves state-of-the-art performance in underwater image restoration tasks, both quantitatively and qualitatively. Our results show that well-designed pure CNN architectures can effectively compete with more complex models, offering a balance between performance and computational efficiency. This work provides new insights into the potential of CNN-based approaches for challenging image restoration tasks in underwater environments. The code is available at \href{https://github.com/CXH-Research/UIR-PolyKernel}{https://github.com/CXH-Research/UIR-PolyKernel}.
High-Fidelity Document Stain Removal via A Large-Scale Real-World Dataset and A Memory-Augmented Transformer
Oct 30, 2024
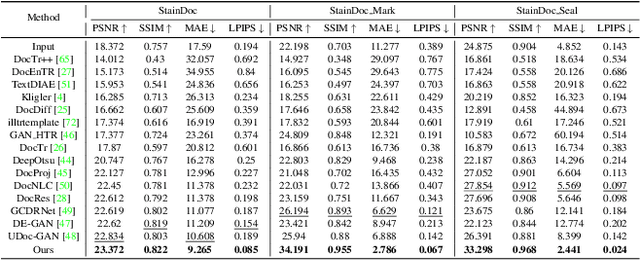

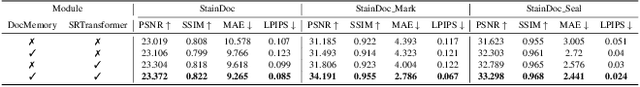
Abstract:Document images are often degraded by various stains, significantly impacting their readability and hindering downstream applications such as document digitization and analysis. The absence of a comprehensive stained document dataset has limited the effectiveness of existing document enhancement methods in removing stains while preserving fine-grained details. To address this challenge, we construct StainDoc, the first large-scale, high-resolution ($2145\times2245$) dataset specifically designed for document stain removal. StainDoc comprises over 5,000 pairs of stained and clean document images across multiple scenes. This dataset encompasses a diverse range of stain types, severities, and document backgrounds, facilitating robust training and evaluation of document stain removal algorithms. Furthermore, we propose StainRestorer, a Transformer-based document stain removal approach. StainRestorer employs a memory-augmented Transformer architecture that captures hierarchical stain representations at part, instance, and semantic levels via the DocMemory module. The Stain Removal Transformer (SRTransformer) leverages these feature representations through a dual attention mechanism: an enhanced spatial attention with an expanded receptive field, and a channel attention captures channel-wise feature importance. This combination enables precise stain removal while preserving document content integrity. Extensive experiments demonstrate StainRestorer's superior performance over state-of-the-art methods on the StainDoc dataset and its variants StainDoc\_Mark and StainDoc\_Seal, establishing a new benchmark for document stain removal. Our work highlights the potential of memory-augmented Transformers for this task and contributes a valuable dataset to advance future research.
Test-Time Intensity Consistency Adaptation for Shadow Detection
Oct 10, 2024
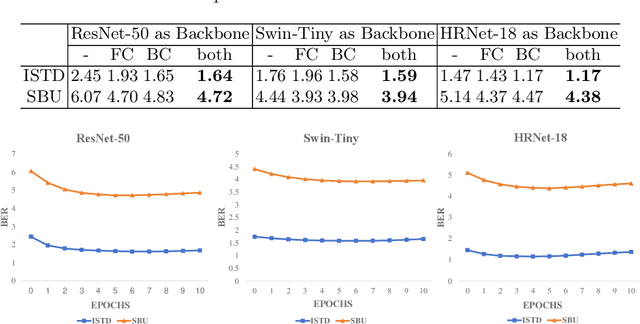
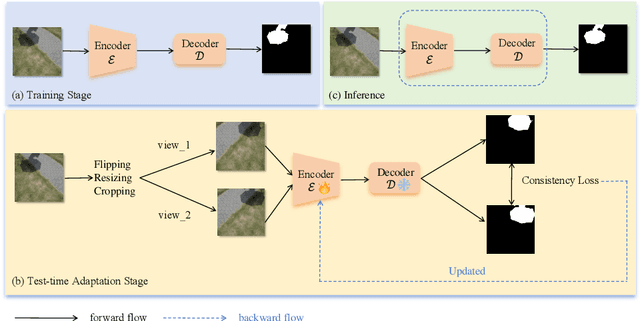
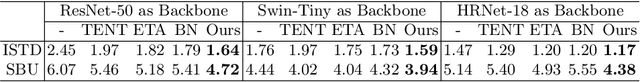
Abstract:Shadow detection is crucial for accurate scene understanding in computer vision, yet it is challenged by the diverse appearances of shadows caused by variations in illumination, object geometry, and scene context. Deep learning models often struggle to generalize to real-world images due to the limited size and diversity of training datasets. To address this, we introduce TICA, a novel framework that leverages light-intensity information during test-time adaptation to enhance shadow detection accuracy. TICA exploits the inherent inconsistencies in light intensity across shadow regions to guide the model toward a more consistent prediction. A basic encoder-decoder model is initially trained on a labeled dataset for shadow detection. Then, during the testing phase, the network is adjusted for each test sample by enforcing consistent intensity predictions between two augmented input image versions. This consistency training specifically targets both foreground and background intersection regions to identify shadow regions within images accurately for robust adaptation. Extensive evaluations on the ISTD and SBU shadow detection datasets reveal that TICA significantly demonstrates that TICA outperforms existing state-of-the-art methods, achieving superior results in balanced error rate (BER).
Enhanced Self-supervised Learning for Multi-modality MRI Segmentation and Classification: A Novel Approach Avoiding Model Collapse
Jul 15, 2024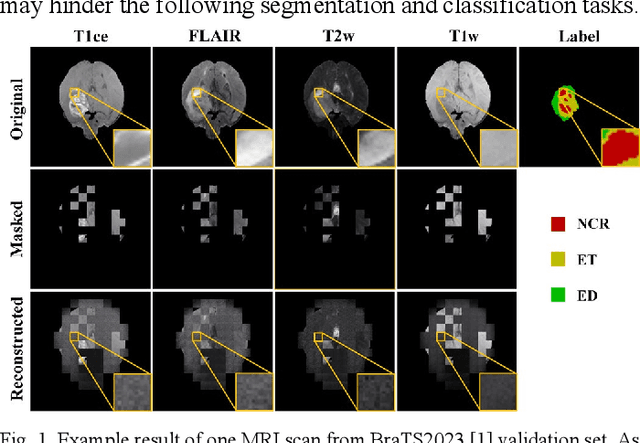
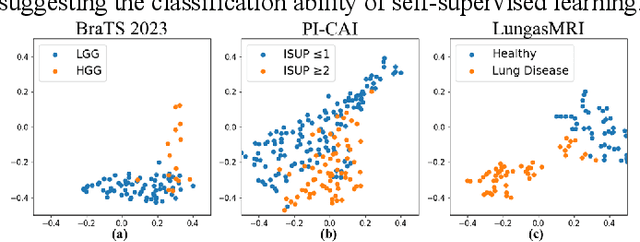
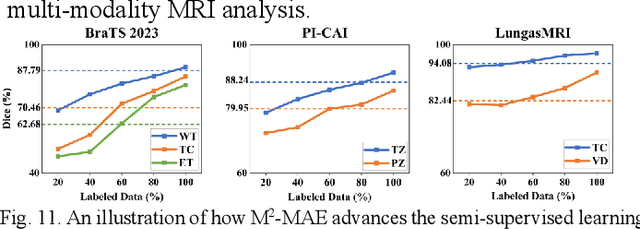
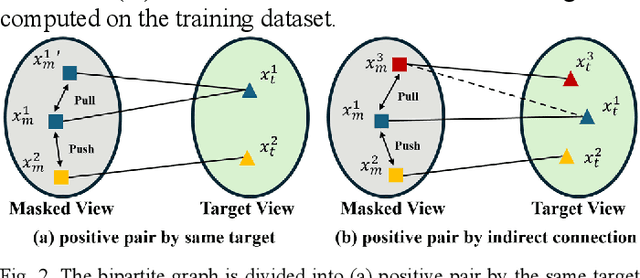
Abstract:Multi-modality magnetic resonance imaging (MRI) can provide complementary information for computer-aided diagnosis. Traditional deep learning algorithms are suitable for identifying specific anatomical structures segmenting lesions and classifying diseases with magnetic resonance images. However, manual labels are limited due to high expense, which hinders further improvement of model accuracy. Self-supervised learning (SSL) can effectively learn feature representations from unlabeled data by pre-training and is demonstrated to be effective in natural image analysis. Most SSL methods ignore the similarity of multi-modality MRI, leading to model collapse. This limits the efficiency of pre-training, causing low accuracy in downstream segmentation and classification tasks. To solve this challenge, we establish and validate a multi-modality MRI masked autoencoder consisting of hybrid mask pattern (HMP) and pyramid barlow twin (PBT) module for SSL on multi-modality MRI analysis. The HMP concatenates three masking steps forcing the SSL to learn the semantic connections of multi-modality images by reconstructing the masking patches. We have proved that the proposed HMP can avoid model collapse. The PBT module exploits the pyramidal hierarchy of the network to construct barlow twin loss between masked and original views, aligning the semantic representations of image patches at different vision scales in latent space. Experiments on BraTS2023, PI-CAI, and lung gas MRI datasets further demonstrate the superiority of our framework over the state-of-the-art. The performance of the segmentation and classification is substantially enhanced, supporting the accurate detection of small lesion areas. The code is available at https://github.com/LinxuanHan/M2-MAE.
Full Bayesian Significance Testing for Neural Networks
Jan 24, 2024



Abstract:Significance testing aims to determine whether a proposition about the population distribution is the truth or not given observations. However, traditional significance testing often needs to derive the distribution of the testing statistic, failing to deal with complex nonlinear relationships. In this paper, we propose to conduct Full Bayesian Significance Testing for neural networks, called \textit{n}FBST, to overcome the limitation in relationship characterization of traditional approaches. A Bayesian neural network is utilized to fit the nonlinear and multi-dimensional relationships with small errors and avoid hard theoretical derivation by computing the evidence value. Besides, \textit{n}FBST can test not only global significance but also local and instance-wise significance, which previous testing methods don't focus on. Moreover, \textit{n}FBST is a general framework that can be extended based on the measures selected, such as Grad-\textit{n}FBST, LRP-\textit{n}FBST, DeepLIFT-\textit{n}FBST, LIME-\textit{n}FBST. A range of experiments on both simulated and real data are conducted to show the advantages of our method.
Encoding Enhanced Complex CNN for Accurate and Highly Accelerated MRI
Jun 21, 2023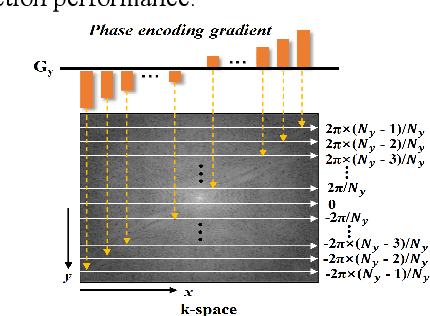
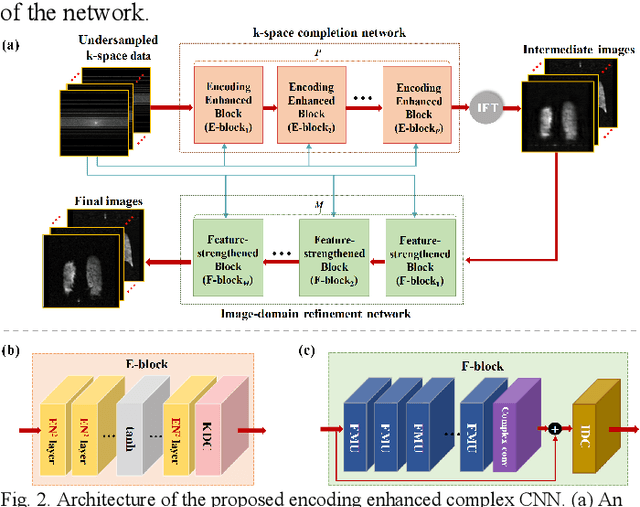
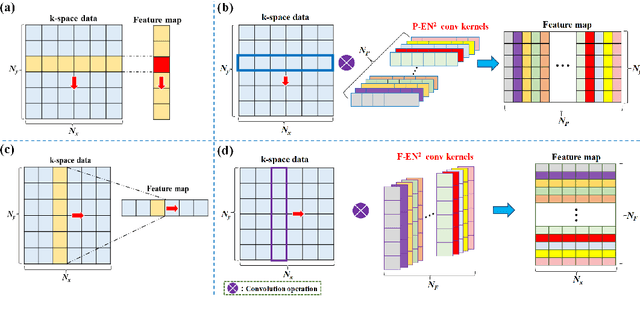
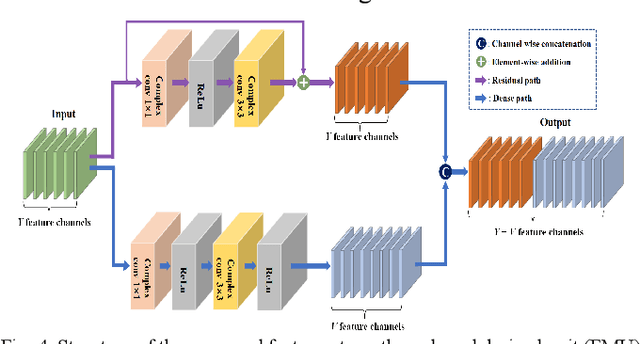
Abstract:Magnetic resonance imaging (MRI) using hyperpolarized noble gases provides a way to visualize the structure and function of human lung, but the long imaging time limits its broad research and clinical applications. Deep learning has demonstrated great potential for accelerating MRI by reconstructing images from undersampled data. However, most existing deep conventional neural networks (CNN) directly apply square convolution to k-space data without considering the inherent properties of k-space sampling, limiting k-space learning efficiency and image reconstruction quality. In this work, we propose an encoding enhanced (EN2) complex CNN for highly undersampled pulmonary MRI reconstruction. EN2 employs convolution along either the frequency or phase-encoding direction, resembling the mechanisms of k-space sampling, to maximize the utilization of the encoding correlation and integrity within a row or column of k-space. We also employ complex convolution to learn rich representations from the complex k-space data. In addition, we develop a feature-strengthened modularized unit to further boost the reconstruction performance. Experiments demonstrate that our approach can accurately reconstruct hyperpolarized 129Xe and 1H lung MRI from 6-fold undersampled k-space data and provide lung function measurements with minimal biases compared with fully-sampled image. These results demonstrate the effectiveness of the proposed algorithmic components and indicate that the proposed approach could be used for accelerated pulmonary MRI in research and clinical lung disease patient care.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge