Yushan Zheng
Promptable Representation Distribution Learning and Data Augmentation for Gigapixel Histopathology WSI Analysis
Dec 19, 2024Abstract:Gigapixel image analysis, particularly for whole slide images (WSIs), often relies on multiple instance learning (MIL). Under the paradigm of MIL, patch image representations are extracted and then fixed during the training of the MIL classifiers for efficiency consideration. However, the invariance of representations makes it difficult to perform data augmentation for WSI-level model training, which significantly limits the performance of the downstream WSI analysis. The current data augmentation methods for gigapixel images either introduce additional computational costs or result in a loss of semantic information, which is hard to meet the requirements for efficiency and stability needed for WSI model training. In this paper, we propose a Promptable Representation Distribution Learning framework (PRDL) for both patch-level representation learning and WSI-level data augmentation. Meanwhile, we explore the use of prompts to guide data augmentation in feature space, which achieves promptable data augmentation for training robust WSI-level models. The experimental results have demonstrated that the proposed method stably outperforms state-of-the-art methods.
Slide-based Graph Collaborative Training for Histopathology Whole Slide Image Analysis
Oct 14, 2024Abstract:The development of computational pathology lies in the consensus that pathological characteristics of tumors are significant guidance for cancer diagnostics. Most existing research focuses on the inner-contextual information within each WSI yet ignores the possible inter-correlations between slides. As the development of tumors is a continuous process involving a series of histological, morphological, and genetic changes that accumulate over time, the similarities and differences between WSIs across various stages, grades, locations and patients should potentially contribute to the representation of WSIs and deserve to be taken into account in WSI modeling. To verify the advancement of introducing the slide inter-correlations into the representation learning of WSIs, we proposed a generic WSI analysis pipeline SlideGCD that can be adapted to any existing Multiple Instance Learning (MIL) frameworks and improve their performance. With the new paradigm, the prior knowledge of cancer development can participate in the end-to-end workflow, which concurrently initializes and refines the slide representation, as a guide for message passing in the slide-based graph. Extensive comparisons and experiments are conducted to validate the effectiveness and robustness of the proposed pipeline across 4 different tasks, including cancer subtyping, cancer staging, survival prediction, and gene mutation prediction, with 7 representative SOTA WSI analysis frameworks as backbones.
SlideGCD: Slide-based Graph Collaborative Training with Knowledge Distillation for Whole Slide Image Classification
Jul 12, 2024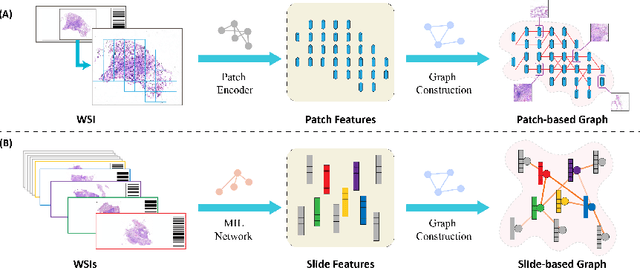
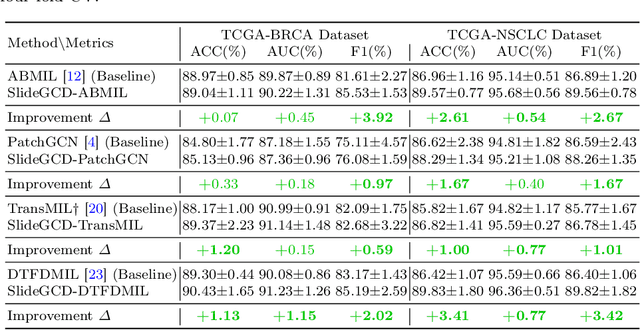
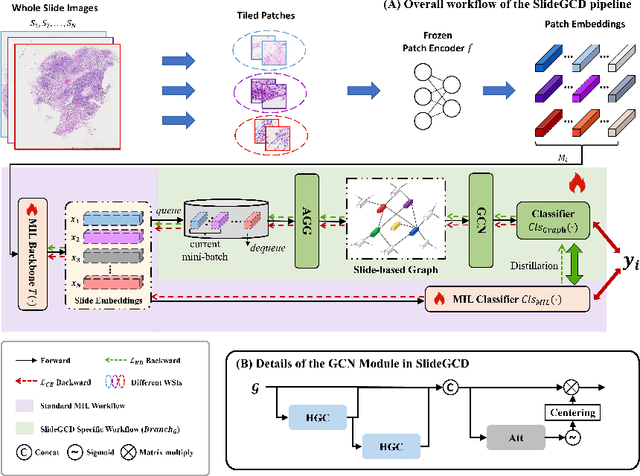
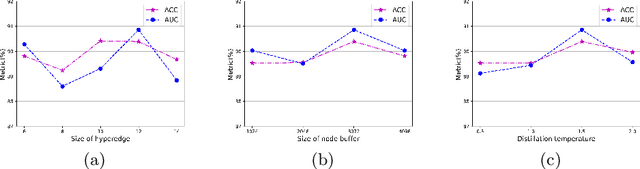
Abstract:Existing WSI analysis methods lie on the consensus that histopathological characteristics of tumors are significant guidance for cancer diagnostics. Particularly, as the evolution of cancers is a continuous process, the correlations and differences across various stages, anatomical locations and patients should be taken into account. However, recent research mainly focuses on the inner-contextual information in a single WSI, ignoring the correlations between slides. To verify whether introducing the slide inter-correlations can bring improvements to WSI representation learning, we propose a generic WSI analysis pipeline SlideGCD that considers the existing multi-instance learning (MIL) methods as the backbone and forge the WSI classification task as a node classification problem. More specifically, SlideGCD declares a node buffer that stores previous slide embeddings for subsequent extensive slide-based graph construction and conducts graph learning to explore the inter-correlations implied in the slide-based graph. Moreover, we frame the MIL classifier and graph learning into two parallel workflows and deploy the knowledge distillation to transfer the differentiable information to the graph neural network. The consistent performance boosting, brought by SlideGCD, of four previous state-of-the-art MIL methods is observed on two TCGA benchmark datasets. The code is available at https://github.com/HFUT-miaLab/SlideGCD.
Lifelong Histopathology Whole Slide Image Retrieval via Distance Consistency Rehearsal
Jul 11, 2024Abstract:Content-based histopathological image retrieval (CBHIR) has gained attention in recent years, offering the capability to return histopathology images that are content-wise similar to the query one from an established database. However, in clinical practice, the continuously expanding size of WSI databases limits the practical application of the current CBHIR methods. In this paper, we propose a Lifelong Whole Slide Retrieval (LWSR) framework to address the challenges of catastrophic forgetting by progressive model updating on continuously growing retrieval database. Our framework aims to achieve the balance between stability and plasticity during continuous learning. To preserve system plasticity, we utilize local memory bank with reservoir sampling method to save instances, which can comprehensively encompass the feature spaces of both old and new tasks. Furthermore, A distance consistency rehearsal (DCR) module is designed to ensure the retrieval queue's consistency for previous tasks, which is regarded as stability within a lifelong CBHIR system. We evaluated the proposed method on four public WSI datasets from TCGA projects. The experimental results have demonstrated the proposed method is effective and is superior to the state-of-the-art methods.
Pan-cancer Histopathology WSI Pre-training with Position-aware Masked Autoencoder
Jul 10, 2024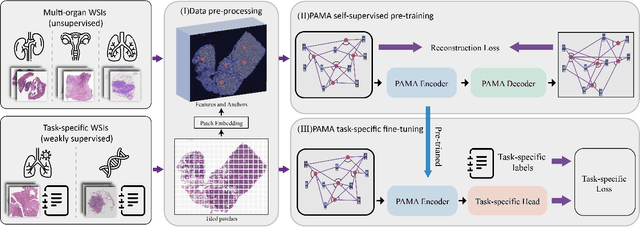
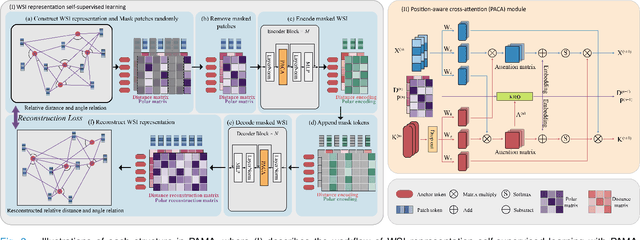
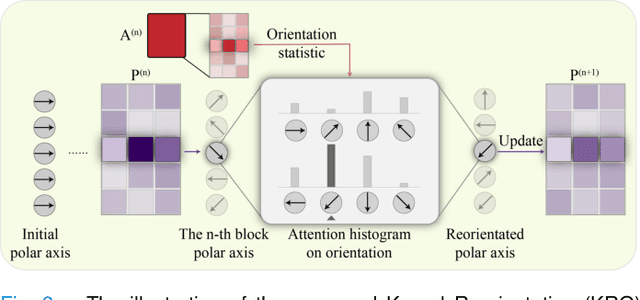
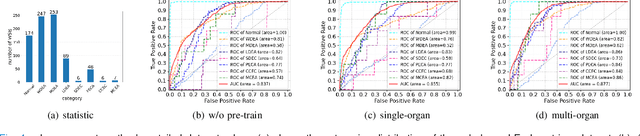
Abstract:Large-scale pre-training models have promoted the development of histopathology image analysis. However, existing self-supervised methods for histopathology images focus on learning patch features, while there is still a lack of available pre-training models for WSI-level feature learning. In this paper, we propose a novel self-supervised learning framework for pan-cancer WSI-level representation pre-training with the designed position-aware masked autoencoder (PAMA). Meanwhile, we propose the position-aware cross-attention (PACA) module with a kernel reorientation (KRO) strategy and an anchor dropout (AD) mechanism. The KRO strategy can capture the complete semantic structure and eliminate ambiguity in WSIs, and the AD contributes to enhancing the robustness and generalization of the model. We evaluated our method on 6 large-scale datasets from multiple organs for pan-cancer classification tasks. The results have demonstrated the effectiveness of PAMA in generalized and discriminative WSI representation learning and pan-cancer WSI pre-training. The proposed method was also compared with \R{7} WSI analysis methods. The experimental results have indicated that our proposed PAMA is superior to the state-of-the-art methods.The code and checkpoints are available at https://github.com/WkEEn/PAMA.
Kernel Attention Transformer (KAT) for Histopathology Whole Slide Image Classification
Jun 27, 2022



Abstract:Transformer has been widely used in histopathology whole slide image (WSI) classification for the purpose of tumor grading, prognosis analysis, etc. However, the design of token-wise self-attention and positional embedding strategy in the common Transformer limits the effectiveness and efficiency in the application to gigapixel histopathology images. In this paper, we propose a kernel attention Transformer (KAT) for histopathology WSI classification. The information transmission of the tokens is achieved by cross-attention between the tokens and a set of kernels related to a set of positional anchors on the WSI. Compared to the common Transformer structure, the proposed KAT can better describe the hierarchical context information of the local regions of the WSI and meanwhile maintains a lower computational complexity. The proposed method was evaluated on a gastric dataset with 2040 WSIs and an endometrial dataset with 2560 WSIs, and was compared with 6 state-of-the-art methods. The experimental results have demonstrated the proposed KAT is effective and efficient in the task of histopathology WSI classification and is superior to the state-of-the-art methods. The code is available at https://github.com/zhengyushan/kat.
Lesion-Aware Contrastive Representation Learning for Histopathology Whole Slide Images Analysis
Jun 27, 2022



Abstract:Local representation learning has been a key challenge to promote the performance of the histopathological whole slide images analysis. The previous representation learning methods followed the supervised learning paradigm. However, manual annotation for large-scale WSIs is time-consuming and labor-intensive. Hence, the self-supervised contrastive learning has recently attracted intensive attention. The present contrastive learning methods treat each sample as a single class, which suffers from class collision problems, especially in the domain of histopathology image analysis. In this paper, we proposed a novel contrastive representation learning framework named Lesion-Aware Contrastive Learning (LACL) for histopathology whole slide image analysis. We built a lesion queue based on the memory bank structure to store the representations of different classes of WSIs, which allowed the contrastive model to selectively define the negative pairs during the training. Moreover, We designed a queue refinement strategy to purify the representations stored in the lesion queue. The experimental results demonstrate that LACL achieves the best performance in histopathology image representation learning on different datasets, and outperforms state-of-the-art methods under different WSI classification benchmarks. The code is available at https://github.com/junl21/lacl.
Histopathology WSI Encoding based on GCNs for Scalable and Efficient Retrieval of Diagnostically Relevant Regions
Apr 16, 2021



Abstract:Content-based histopathological image retrieval (CBHIR) has become popular in recent years in the domain of histopathological image analysis. CBHIR systems provide auxiliary diagnosis information for pathologists by searching for and returning regions that are contently similar to the region of interest (ROI) from a pre-established database. While, it is challenging and yet significant in clinical applications to retrieve diagnostically relevant regions from a database that consists of histopathological whole slide images (WSIs) for a query ROI. In this paper, we propose a novel framework for regions retrieval from WSI-database based on hierarchical graph convolutional networks (GCNs) and Hash technique. Compared to the present CBHIR framework, the structural information of WSI is preserved through graph embedding of GCNs, which makes the retrieval framework more sensitive to regions that are similar in tissue distribution. Moreover, benefited from the hierarchical GCN structures, the proposed framework has good scalability for both the size and shape variation of ROIs. It allows the pathologist defining query regions using free curves according to the appearance of tissue. Thirdly, the retrieval is achieved based on Hash technique, which ensures the framework is efficient and thereby adequate for practical large-scale WSI-database. The proposed method was validated on two public datasets for histopathological WSI analysis and compared to the state-of-the-art methods. The proposed method achieved mean average precision above 0.857 on the ACDC-LungHP dataset and above 0.864 on the Camelyon16 dataset in the irregular region retrieval tasks, which are superior to the state-of-the-art methods. The average retrieval time from a database within 120 WSIs is 0.802 ms.
An Optimized H.266/VVC Software Decoder On Mobile Platform
Mar 05, 2021



Abstract:As the successor of H.265/HEVC, the new versatile video coding standard (H.266/VVC) can provide up to 50% bitrate saving with the same subjective quality, at the cost of increased decoding complexity. To accelerate the application of the new coding standard, a real-time H.266/VVC software decoder that can support various platforms is implemented, where SIMD technologies, parallelism optimization, and the acceleration strategies based on the characteristics of each coding tool are applied. As the mobile devices have become an essential carrier for video services nowadays, the mentioned optimization efforts are not only implemented for the x86 platform, but more importantly utilized to highly optimize the decoding performance on the ARM platform in this work. The experimental results show that when running on the Apple A14 SoC (iPhone 12pro), the average single-thread decoding speed of the present implementation can achieve 53fps (RA and LB) for full HD (1080p) bitstreams generated by VTM-11.0 reference software using 8bit Common Test Conditions (CTC). When multi-threading is enabled, an average of 32 fps (RA) can be achieved when decoding the 4K bitstreams.
Deep Learning Methods for Lung Cancer Segmentation in Whole-slide Histopathology Images -- the ACDC@LungHP Challenge 2019
Aug 21, 2020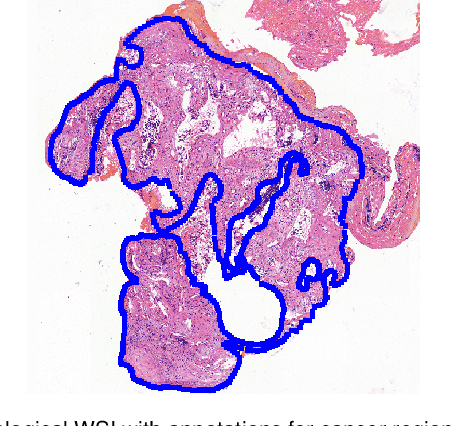
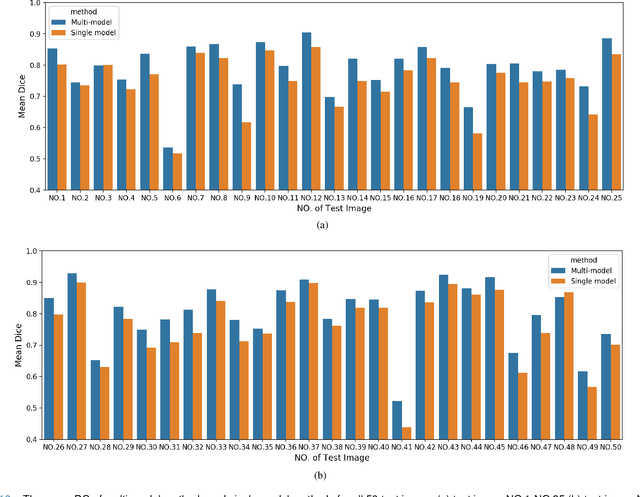
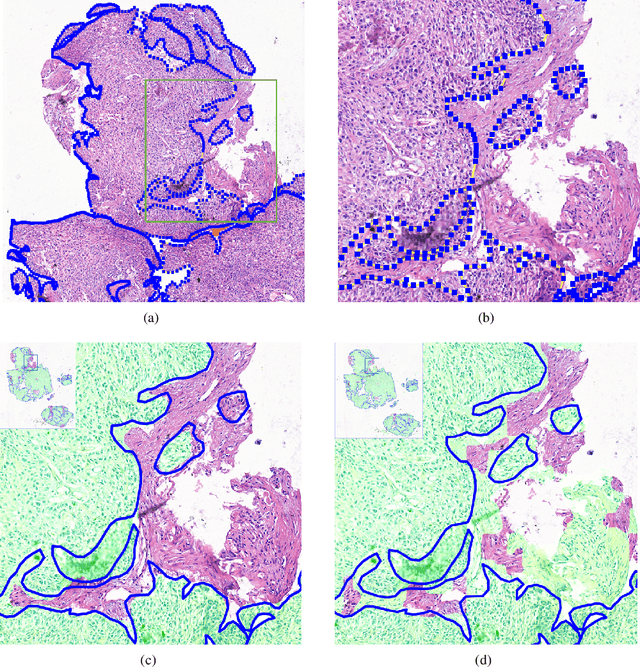
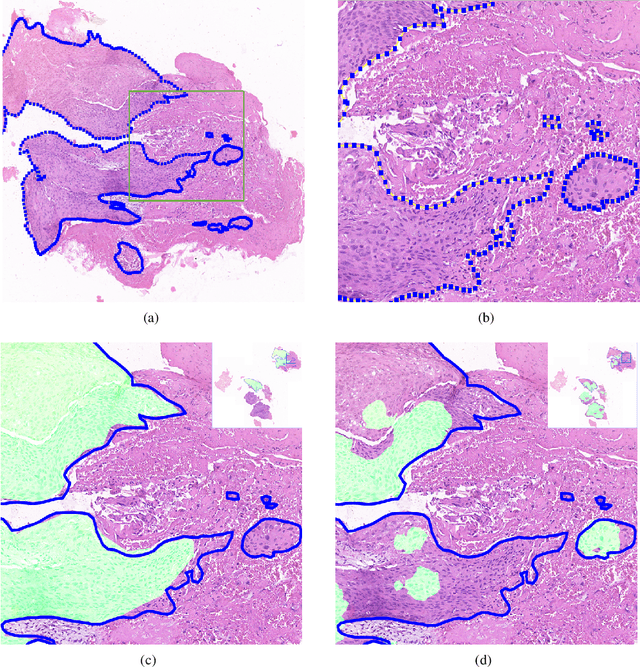
Abstract:Accurate segmentation of lung cancer in pathology slides is a critical step in improving patient care. We proposed the ACDC@LungHP (Automatic Cancer Detection and Classification in Whole-slide Lung Histopathology) challenge for evaluating different computer-aided diagnosis (CADs) methods on the automatic diagnosis of lung cancer. The ACDC@LungHP 2019 focused on segmentation (pixel-wise detection) of cancer tissue in whole slide imaging (WSI), using an annotated dataset of 150 training images and 50 test images from 200 patients. This paper reviews this challenge and summarizes the top 10 submitted methods for lung cancer segmentation. All methods were evaluated using the false positive rate, false negative rate, and DICE coefficient (DC). The DC ranged from 0.7354$\pm$0.1149 to 0.8372$\pm$0.0858. The DC of the best method was close to the inter-observer agreement (0.8398$\pm$0.0890). All methods were based on deep learning and categorized into two groups: multi-model method and single model method. In general, multi-model methods were significantly better ($\textit{p}$<$0.01$) than single model methods, with mean DC of 0.7966 and 0.7544, respectively. Deep learning based methods could potentially help pathologists find suspicious regions for further analysis of lung cancer in WSI.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge