Fengying Xie
Rectification Reimagined: A Unified Mamba Model for Image Correction and Rectangling with Prompts
Dec 21, 2025



Abstract:Image correction and rectangling are valuable tasks in practical photography systems such as smartphones. Recent remarkable advancements in deep learning have undeniably brought about substantial performance improvements in these fields. Nevertheless, existing methods mainly rely on task-specific architectures. This significantly restricts their generalization ability and effective application across a wide range of different tasks. In this paper, we introduce the Unified Rectification Framework (UniRect), a comprehensive approach that addresses these practical tasks from a consistent distortion rectification perspective. Our approach incorporates various task-specific inverse problems into a general distortion model by simulating different types of lenses. To handle diverse distortions, UniRect adopts one task-agnostic rectification framework with a dual-component structure: a {Deformation Module}, which utilizes a novel Residual Progressive Thin-Plate Spline (RP-TPS) model to address complex geometric deformations, and a subsequent Restoration Module, which employs Residual Mamba Blocks (RMBs) to counteract the degradation caused by the deformation process and enhance the fidelity of the output image. Moreover, a Sparse Mixture-of-Experts (SMoEs) structure is designed to circumvent heavy task competition in multi-task learning due to varying distortions. Extensive experiments demonstrate that our models have achieved state-of-the-art performance compared with other up-to-date methods.
Adapt CLIP as Aggregation Instructor for Image Dehazing
Aug 22, 2024



Abstract:Most dehazing methods suffer from limited receptive field and do not explore the rich semantic prior encapsulated in vision-language models, which have proven effective in downstream tasks. In this paper, we introduce CLIPHaze, a pioneering hybrid framework that synergizes the efficient global modeling of Mamba with the prior knowledge and zero-shot capabilities of CLIP to address both issues simultaneously. Specifically, our method employs parallel state space model and window-based self-attention to obtain global contextual dependency and local fine-grained perception, respectively. To seamlessly aggregate information from both paths, we introduce CLIP-instructed Aggregation Module (CAM). For non-homogeneous and homogeneous haze, CAM leverages zero-shot estimated haze density map and high-quality image embedding without degradation information to explicitly and implicitly determine the optimal neural operation range for each pixel, thereby adaptively fusing two paths with different receptive fields. Extensive experiments on various benchmarks demonstrate that CLIPHaze achieves state-of-the-art (SOTA) performance, particularly in non-homogeneous haze. Code will be publicly after acceptance.
Pan-cancer Histopathology WSI Pre-training with Position-aware Masked Autoencoder
Jul 10, 2024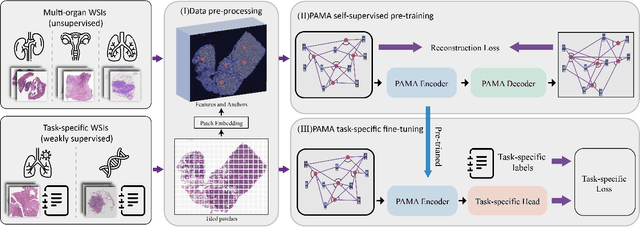
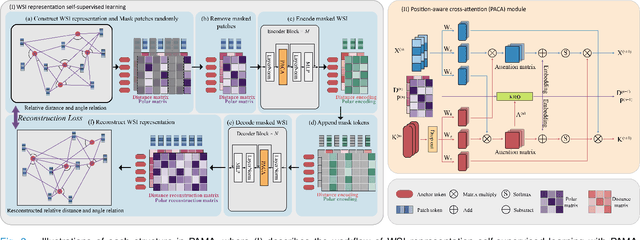
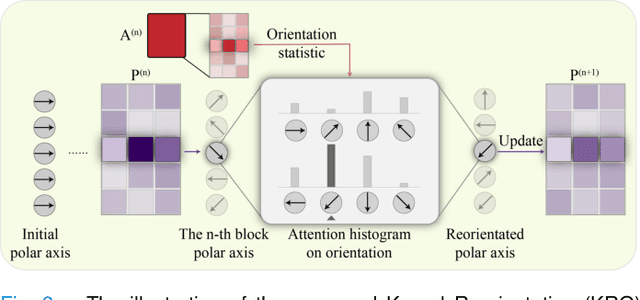
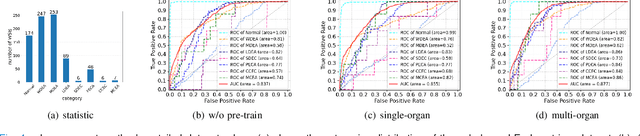
Abstract:Large-scale pre-training models have promoted the development of histopathology image analysis. However, existing self-supervised methods for histopathology images focus on learning patch features, while there is still a lack of available pre-training models for WSI-level feature learning. In this paper, we propose a novel self-supervised learning framework for pan-cancer WSI-level representation pre-training with the designed position-aware masked autoencoder (PAMA). Meanwhile, we propose the position-aware cross-attention (PACA) module with a kernel reorientation (KRO) strategy and an anchor dropout (AD) mechanism. The KRO strategy can capture the complete semantic structure and eliminate ambiguity in WSIs, and the AD contributes to enhancing the robustness and generalization of the model. We evaluated our method on 6 large-scale datasets from multiple organs for pan-cancer classification tasks. The results have demonstrated the effectiveness of PAMA in generalized and discriminative WSI representation learning and pan-cancer WSI pre-training. The proposed method was also compared with \R{7} WSI analysis methods. The experimental results have indicated that our proposed PAMA is superior to the state-of-the-art methods.The code and checkpoints are available at https://github.com/WkEEn/PAMA.
Prototypical Information Bottlenecking and Disentangling for Multimodal Cancer Survival Prediction
Jan 03, 2024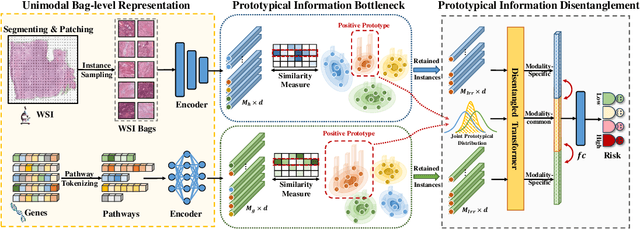
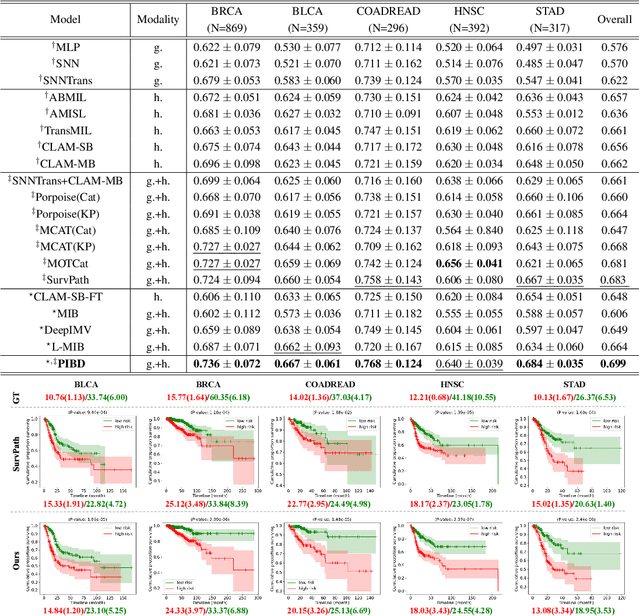
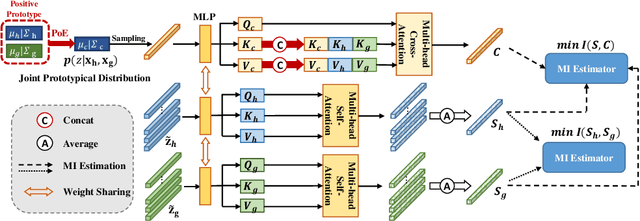
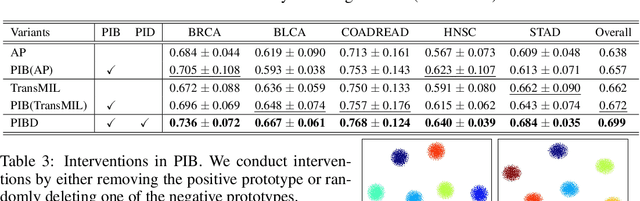
Abstract:Multimodal learning significantly benefits cancer survival prediction, especially the integration of pathological images and genomic data. Despite advantages of multimodal learning for cancer survival prediction, massive redundancy in multimodal data prevents it from extracting discriminative and compact information: (1) An extensive amount of intra-modal task-unrelated information blurs discriminability, especially for gigapixel whole slide images (WSIs) with many patches in pathology and thousands of pathways in genomic data, leading to an ``intra-modal redundancy" issue. (2) Duplicated information among modalities dominates the representation of multimodal data, which makes modality-specific information prone to being ignored, resulting in an ``inter-modal redundancy" issue. To address these, we propose a new framework, Prototypical Information Bottlenecking and Disentangling (PIBD), consisting of Prototypical Information Bottleneck (PIB) module for intra-modal redundancy and Prototypical Information Disentanglement (PID) module for inter-modal redundancy. Specifically, a variant of information bottleneck, PIB, is proposed to model prototypes approximating a bunch of instances for different risk levels, which can be used for selection of discriminative instances within modality. PID module decouples entangled multimodal data into compact distinct components: modality-common and modality-specific knowledge, under the guidance of the joint prototypical distribution. Extensive experiments on five cancer benchmark datasets demonstrated our superiority over other methods.
ECL: Class-Enhancement Contrastive Learning for Long-tailed Skin Lesion Classification
Jul 09, 2023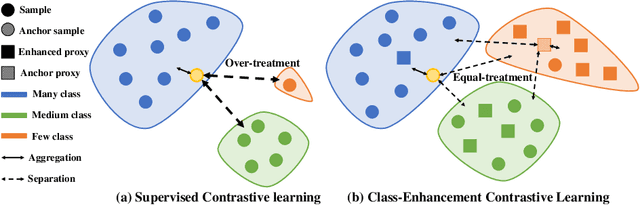
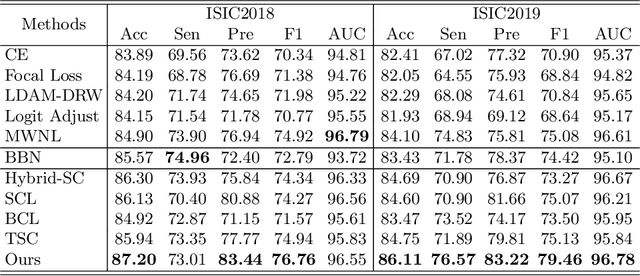
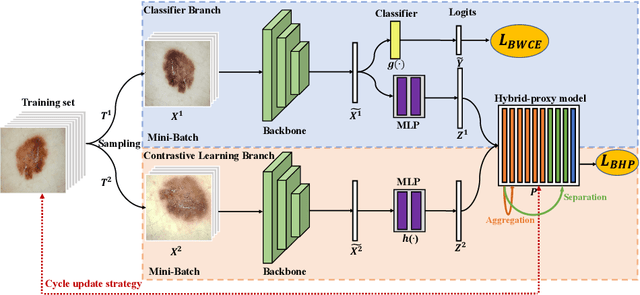
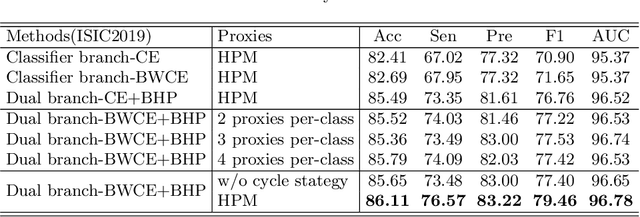
Abstract:Skin image datasets often suffer from imbalanced data distribution, exacerbating the difficulty of computer-aided skin disease diagnosis. Some recent works exploit supervised contrastive learning (SCL) for this long-tailed challenge. Despite achieving significant performance, these SCL-based methods focus more on head classes, yet ignoring the utilization of information in tail classes. In this paper, we propose class-Enhancement Contrastive Learning (ECL), which enriches the information of minority classes and treats different classes equally. For information enhancement, we design a hybrid-proxy model to generate class-dependent proxies and propose a cycle update strategy for parameters optimization. A balanced-hybrid-proxy loss is designed to exploit relations between samples and proxies with different classes treated equally. Taking both "imbalanced data" and "imbalanced diagnosis difficulty" into account, we further present a balanced-weighted cross-entropy loss following curriculum learning schedule. Experimental results on the classification of imbalanced skin lesion data have demonstrated the superiority and effectiveness of our method.
TFormer: A throughout fusion transformer for multi-modal skin lesion diagnosis
Nov 21, 2022



Abstract:Multi-modal skin lesion diagnosis (MSLD) has achieved remarkable success by modern computer-aided diagnosis technology based on deep convolutions. However, the information aggregation across modalities in MSLD remains challenging due to severity unaligned spatial resolution (dermoscopic image and clinical image) and heterogeneous data (dermoscopic image and patients' meta-data). Limited by the intrinsic local attention, most recent MSLD pipelines using pure convolutions struggle to capture representative features in shallow layers, thus the fusion across different modalities is usually done at the end of the pipelines, even at the last layer, leading to an insufficient information aggregation. To tackle the issue, we introduce a pure transformer-based method, which we refer to as ``Throughout Fusion Transformer (TFormer)", for sufficient information intergration in MSLD. Different from the existing approaches with convolutions, the proposed network leverages transformer as feature extraction backbone, bringing more representative shallow features. We then carefully design a stack of dual-branch hierarchical multi-modal transformer (HMT) blocks to fuse information across different image modalities in a stage-by-stage way. With the aggregated information of image modalities, a multi-modal transformer post-fusion (MTP) block is designed to integrate features across image and non-image data. Such a strategy that information of the image modalities is firstly fused then the heterogeneous ones enables us to better divide and conquer the two major challenges while ensuring inter-modality dynamics are effectively modeled. Experiments conducted on the public Derm7pt dataset validate the superiority of the proposed method. Our TFormer outperforms other state-of-the-art methods. Ablation experiments also suggest the effectiveness of our designs.
A Rotation Meanout Network with Invariance for Dermoscopy Image Classification and Retrieval
Aug 01, 2022



Abstract:The computer-aided diagnosis (CAD) system can provide a reference basis for the clinical diagnosis of skin diseases. Convolutional neural networks (CNNs) can not only extract visual elements such as colors and shapes but also semantic features. As such they have made great improvements in many tasks of dermoscopy images. The imaging of dermoscopy has no main direction, indicating that there are a large number of skin lesion target rotations in the datasets. However, CNNs lack anti-rotation ability, which is bound to affect the feature extraction ability of CNNs. We propose a rotation meanout (RM) network to extract rotation invariance features from dermoscopy images. In RM, each set of rotated feature maps corresponds to a set of weight-sharing convolution outputs and they are fused using meanout operation to obtain the final feature maps. Through theoretical derivation, the proposed RM network is rotation-equivariant and can extract rotation-invariant features when being followed by the global average pooling (GAP) operation. The extracted rotation-invariant features can better represent the original data in classification and retrieval tasks for dermoscopy images. The proposed RM is a general operation, which does not change the network structure or increase any parameter, and can be flexibly embedded in any part of CNNs. Extensive experiments are conducted on a dermoscopy image dataset. The results show our method outperforms other anti-rotation methods and achieves great improvements in dermoscopy image classification and retrieval tasks, indicating the potential of rotation invariance in the field of dermoscopy images.
Kernel Attention Transformer (KAT) for Histopathology Whole Slide Image Classification
Jun 27, 2022



Abstract:Transformer has been widely used in histopathology whole slide image (WSI) classification for the purpose of tumor grading, prognosis analysis, etc. However, the design of token-wise self-attention and positional embedding strategy in the common Transformer limits the effectiveness and efficiency in the application to gigapixel histopathology images. In this paper, we propose a kernel attention Transformer (KAT) for histopathology WSI classification. The information transmission of the tokens is achieved by cross-attention between the tokens and a set of kernels related to a set of positional anchors on the WSI. Compared to the common Transformer structure, the proposed KAT can better describe the hierarchical context information of the local regions of the WSI and meanwhile maintains a lower computational complexity. The proposed method was evaluated on a gastric dataset with 2040 WSIs and an endometrial dataset with 2560 WSIs, and was compared with 6 state-of-the-art methods. The experimental results have demonstrated the proposed KAT is effective and efficient in the task of histopathology WSI classification and is superior to the state-of-the-art methods. The code is available at https://github.com/zhengyushan/kat.
Lesion-Aware Contrastive Representation Learning for Histopathology Whole Slide Images Analysis
Jun 27, 2022



Abstract:Local representation learning has been a key challenge to promote the performance of the histopathological whole slide images analysis. The previous representation learning methods followed the supervised learning paradigm. However, manual annotation for large-scale WSIs is time-consuming and labor-intensive. Hence, the self-supervised contrastive learning has recently attracted intensive attention. The present contrastive learning methods treat each sample as a single class, which suffers from class collision problems, especially in the domain of histopathology image analysis. In this paper, we proposed a novel contrastive representation learning framework named Lesion-Aware Contrastive Learning (LACL) for histopathology whole slide image analysis. We built a lesion queue based on the memory bank structure to store the representations of different classes of WSIs, which allowed the contrastive model to selectively define the negative pairs during the training. Moreover, We designed a queue refinement strategy to purify the representations stored in the lesion queue. The experimental results demonstrate that LACL achieves the best performance in histopathology image representation learning on different datasets, and outperforms state-of-the-art methods under different WSI classification benchmarks. The code is available at https://github.com/junl21/lacl.
Histopathology WSI Encoding based on GCNs for Scalable and Efficient Retrieval of Diagnostically Relevant Regions
Apr 16, 2021



Abstract:Content-based histopathological image retrieval (CBHIR) has become popular in recent years in the domain of histopathological image analysis. CBHIR systems provide auxiliary diagnosis information for pathologists by searching for and returning regions that are contently similar to the region of interest (ROI) from a pre-established database. While, it is challenging and yet significant in clinical applications to retrieve diagnostically relevant regions from a database that consists of histopathological whole slide images (WSIs) for a query ROI. In this paper, we propose a novel framework for regions retrieval from WSI-database based on hierarchical graph convolutional networks (GCNs) and Hash technique. Compared to the present CBHIR framework, the structural information of WSI is preserved through graph embedding of GCNs, which makes the retrieval framework more sensitive to regions that are similar in tissue distribution. Moreover, benefited from the hierarchical GCN structures, the proposed framework has good scalability for both the size and shape variation of ROIs. It allows the pathologist defining query regions using free curves according to the appearance of tissue. Thirdly, the retrieval is achieved based on Hash technique, which ensures the framework is efficient and thereby adequate for practical large-scale WSI-database. The proposed method was validated on two public datasets for histopathological WSI analysis and compared to the state-of-the-art methods. The proposed method achieved mean average precision above 0.857 on the ACDC-LungHP dataset and above 0.864 on the Camelyon16 dataset in the irregular region retrieval tasks, which are superior to the state-of-the-art methods. The average retrieval time from a database within 120 WSIs is 0.802 ms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge