Kunming Tang
Promptable Representation Distribution Learning and Data Augmentation for Gigapixel Histopathology WSI Analysis
Dec 19, 2024Abstract:Gigapixel image analysis, particularly for whole slide images (WSIs), often relies on multiple instance learning (MIL). Under the paradigm of MIL, patch image representations are extracted and then fixed during the training of the MIL classifiers for efficiency consideration. However, the invariance of representations makes it difficult to perform data augmentation for WSI-level model training, which significantly limits the performance of the downstream WSI analysis. The current data augmentation methods for gigapixel images either introduce additional computational costs or result in a loss of semantic information, which is hard to meet the requirements for efficiency and stability needed for WSI model training. In this paper, we propose a Promptable Representation Distribution Learning framework (PRDL) for both patch-level representation learning and WSI-level data augmentation. Meanwhile, we explore the use of prompts to guide data augmentation in feature space, which achieves promptable data augmentation for training robust WSI-level models. The experimental results have demonstrated that the proposed method stably outperforms state-of-the-art methods.
Pan-cancer Histopathology WSI Pre-training with Position-aware Masked Autoencoder
Jul 10, 2024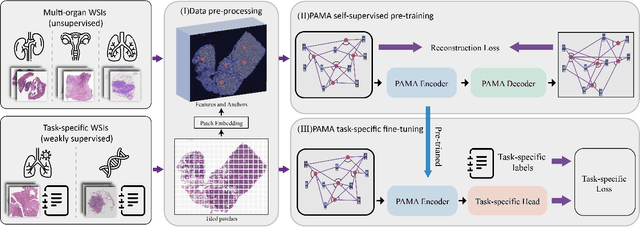
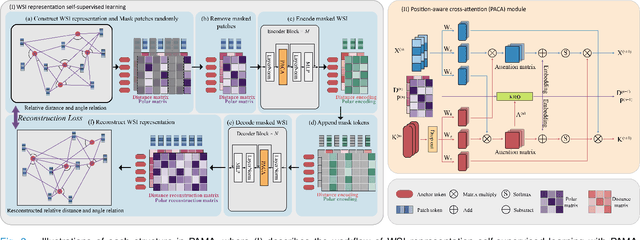
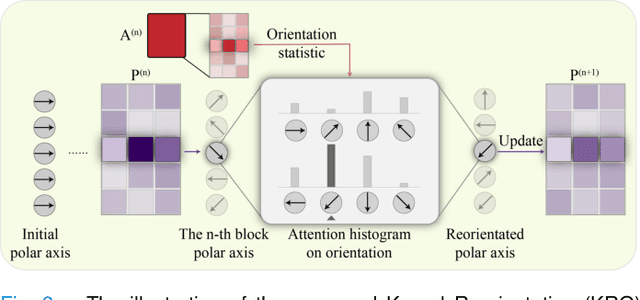
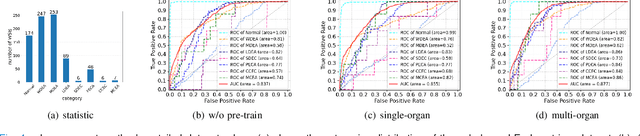
Abstract:Large-scale pre-training models have promoted the development of histopathology image analysis. However, existing self-supervised methods for histopathology images focus on learning patch features, while there is still a lack of available pre-training models for WSI-level feature learning. In this paper, we propose a novel self-supervised learning framework for pan-cancer WSI-level representation pre-training with the designed position-aware masked autoencoder (PAMA). Meanwhile, we propose the position-aware cross-attention (PACA) module with a kernel reorientation (KRO) strategy and an anchor dropout (AD) mechanism. The KRO strategy can capture the complete semantic structure and eliminate ambiguity in WSIs, and the AD contributes to enhancing the robustness and generalization of the model. We evaluated our method on 6 large-scale datasets from multiple organs for pan-cancer classification tasks. The results have demonstrated the effectiveness of PAMA in generalized and discriminative WSI representation learning and pan-cancer WSI pre-training. The proposed method was also compared with \R{7} WSI analysis methods. The experimental results have indicated that our proposed PAMA is superior to the state-of-the-art methods.The code and checkpoints are available at https://github.com/WkEEn/PAMA.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge