Yixiong Liang
Dynamic Residual Encoding with Slide-Level Contrastive Learning for End-to-End Whole Slide Image Representation
Nov 07, 2025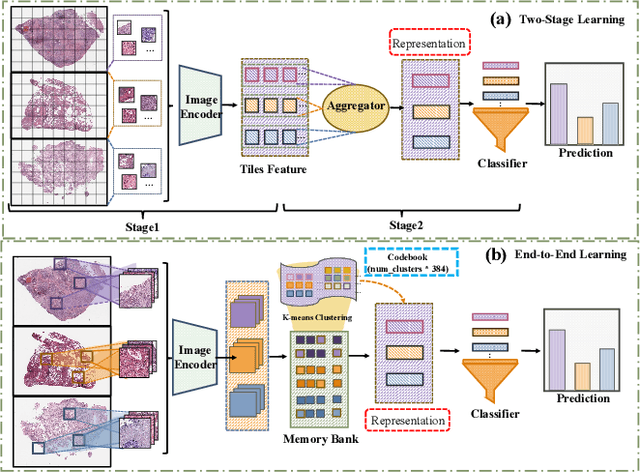
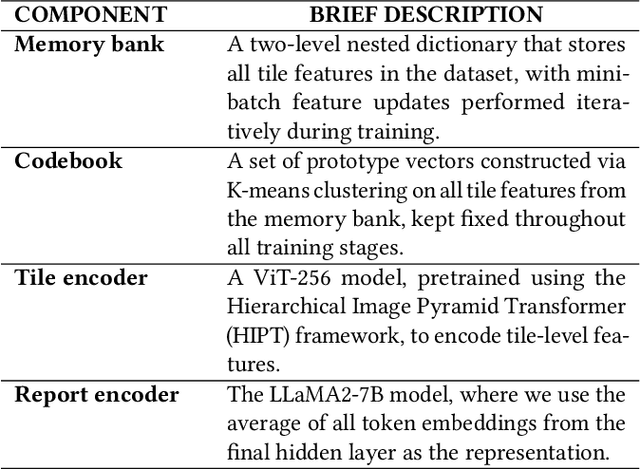
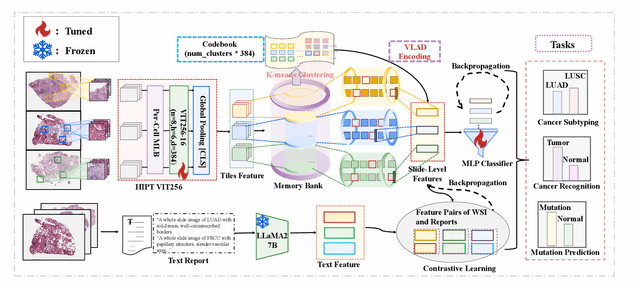
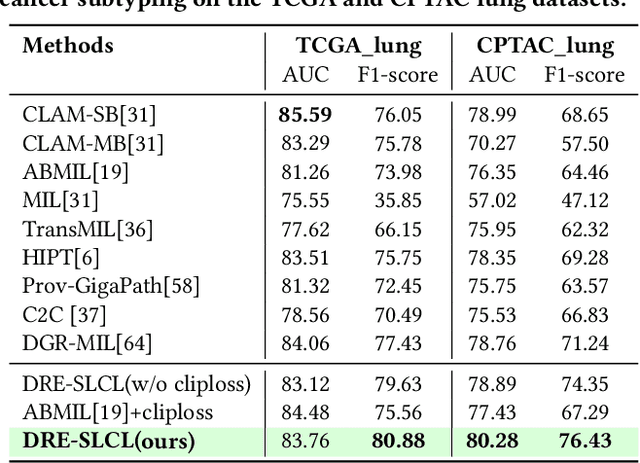
Abstract:Whole Slide Image (WSI) representation is critical for cancer subtyping, cancer recognition and mutation prediction.Training an end-to-end WSI representation model poses significant challenges, as a standard gigapixel slide can contain tens of thousands of image tiles, making it difficult to compute gradients of all tiles in a single mini-batch due to current GPU limitations. To address this challenge, we propose a method of dynamic residual encoding with slide-level contrastive learning (DRE-SLCL) for end-to-end WSI representation. Our approach utilizes a memory bank to store the features of tiles across all WSIs in the dataset. During training, a mini-batch usually contains multiple WSIs. For each WSI in the batch, a subset of tiles is randomly sampled and their features are computed using a tile encoder. Then, additional tile features from the same WSI are selected from the memory bank. The representation of each individual WSI is generated using a residual encoding technique that incorporates both the sampled features and those retrieved from the memory bank. Finally, the slide-level contrastive loss is computed based on the representations and histopathology reports ofthe WSIs within the mini-batch. Experiments conducted over cancer subtyping, cancer recognition, and mutation prediction tasks proved the effectiveness of the proposed DRE-SLCL method.
* 8pages, 3figures, published to ACM Digital Library
HRDecoder: High-Resolution Decoder Network for Fundus Image Lesion Segmentation
Nov 06, 2024Abstract:High resolution is crucial for precise segmentation in fundus images, yet handling high-resolution inputs incurs considerable GPU memory costs, with diminishing performance gains as overhead increases. To address this issue while tackling the challenge of segmenting tiny objects, recent studies have explored local-global fusion methods. These methods preserve fine details using local regions and capture long-range context information from downscaled global images. However, the necessity of multiple forward passes inevitably incurs significant computational overhead, adversely affecting inference speed. In this paper, we propose HRDecoder, a simple High-Resolution Decoder network for fundus lesion segmentation. It integrates a high-resolution representation learning module to capture fine-grained local features and a high-resolution fusion module to fuse multi-scale predictions. Our method effectively improves the overall segmentation accuracy of fundus lesions while consuming reasonable memory and computational overhead, and maintaining satisfying inference speed. Experimental results on the IDRID and DDR datasets demonstrate the effectiveness of our method. Code is available at https://github.com/CVIU-CSU/HRDecoder.
An efficient framework based on large foundation model for cervical cytopathology whole slide image screening
Jul 16, 2024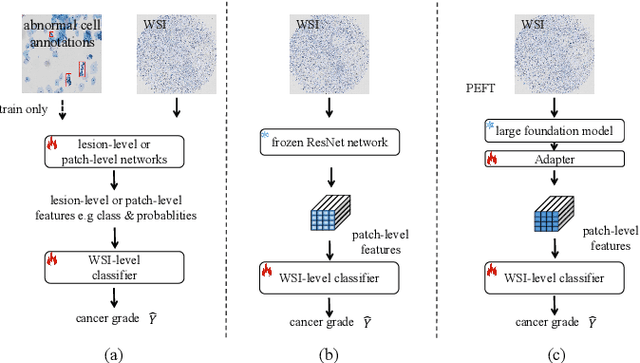
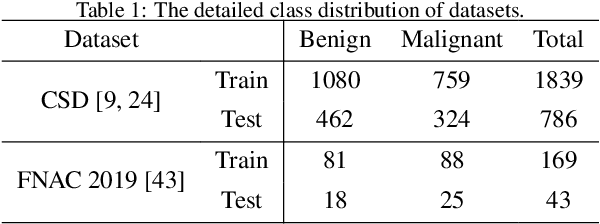
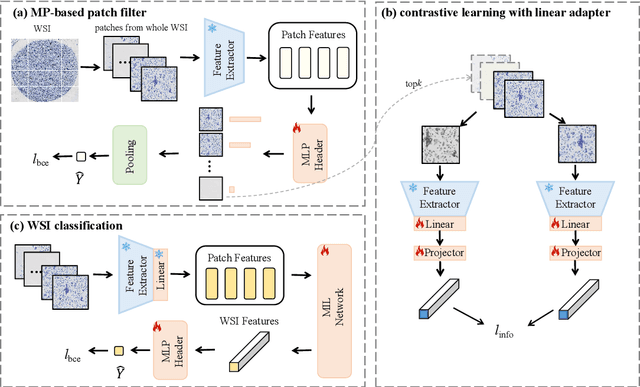
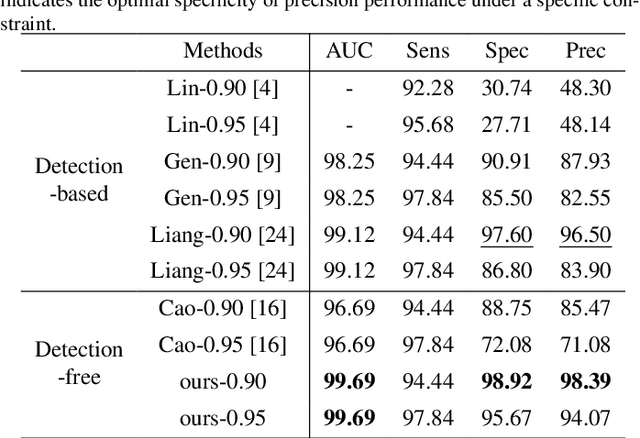
Abstract:Current cervical cytopathology whole slide image (WSI) screening primarily relies on detection-based approaches, which are limited in performance due to the expense and time-consuming annotation process. Multiple Instance Learning (MIL), a weakly supervised approach that relies solely on bag-level labels, can effectively alleviate these challenges. Nonetheless, MIL commonly employs frozen pretrained models or self-supervised learning for feature extraction, which suffers from low efficacy or inefficiency. In this paper, we propose an efficient framework for cervical cytopathology WSI classification using only WSI-level labels through unsupervised and weakly supervised learning. Given the sparse and dispersed nature of abnormal cells within cytopathological WSIs, we propose a strategy that leverages the pretrained foundation model to filter the top$k$ high-risk patches. Subsequently, we suggest parameter-efficient fine-tuning (PEFT) of a large foundation model using contrastive learning on the filtered patches to enhance its representation ability for task-specific signals. By training only the added linear adapters, we enhance the learning of patch-level features with substantially reduced time and memory consumption. Experiments conducted on the CSD and FNAC 2019 datasets demonstrate that the proposed method enhances the performance of various MIL methods and achieves state-of-the-art (SOTA) performance. The code and trained models are publicly available at https://github.com/CVIU-CSU/TCT-InfoNCE.
Unsupervised Collaborative Metric Learning with Mixed-Scale Groups for General Object Retrieval
Mar 16, 2024



Abstract:The task of searching for visual objects in a large image dataset is difficult because it requires efficient matching and accurate localization of objects that can vary in size. Although the segment anything model (SAM) offers a potential solution for extracting object spatial context, learning embeddings for local objects remains a challenging problem. This paper presents a novel unsupervised deep metric learning approach, termed unsupervised collaborative metric learning with mixed-scale groups (MS-UGCML), devised to learn embeddings for objects of varying scales. Following this, a benchmark of challenges is assembled by utilizing COCO 2017 and VOC 2007 datasets to facilitate the training and evaluation of general object retrieval models. Finally, we conduct comprehensive ablation studies and discuss the complexities faced within the domain of general object retrieval. Our object retrieval evaluations span a range of datasets, including BelgaLogos, Visual Genome, LVIS, in addition to a challenging evaluation set that we have individually assembled for open-vocabulary evaluation. These comprehensive evaluations effectively highlight the robustness of our unsupervised MS-UGCML approach, with an object level and image level mAPs improvement of up to 6.69% and 10.03%, respectively. The code is publicly available at https://github.com/dengyuhai/MS-UGCML.
Coded Residual Transform for Generalizable Deep Metric Learning
Oct 09, 2022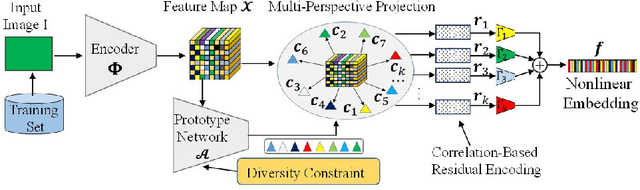

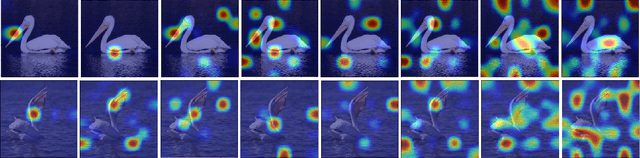
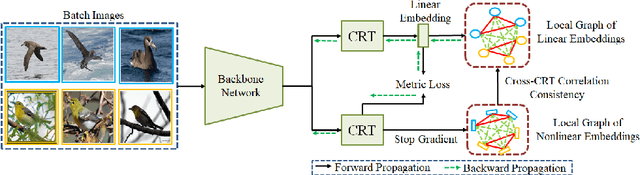
Abstract:A fundamental challenge in deep metric learning is the generalization capability of the feature embedding network model since the embedding network learned on training classes need to be evaluated on new test classes. To address this challenge, in this paper, we introduce a new method called coded residual transform (CRT) for deep metric learning to significantly improve its generalization capability. Specifically, we learn a set of diversified prototype features, project the feature map onto each prototype, and then encode its features using their projection residuals weighted by their correlation coefficients with each prototype. The proposed CRT method has the following two unique characteristics. First, it represents and encodes the feature map from a set of complimentary perspectives based on projections onto diversified prototypes. Second, unlike existing transformer-based feature representation approaches which encode the original values of features based on global correlation analysis, the proposed coded residual transform encodes the relative differences between the original features and their projected prototypes. Embedding space density and spectral decay analysis show that this multi-perspective projection onto diversified prototypes and coded residual representation are able to achieve significantly improved generalization capability in metric learning. Finally, to further enhance the generalization performance, we propose to enforce the consistency on their feature similarity matrices between coded residual transforms with different sizes of projection prototypes and embedding dimensions. Our extensive experimental results and ablation studies demonstrate that the proposed CRT method outperform the state-of-the-art deep metric learning methods by large margins and improving upon the current best method by up to 4.28% on the CUB dataset.
Exploring Contextual Relationships for Cervical Abnormal Cell Detection
Jul 18, 2022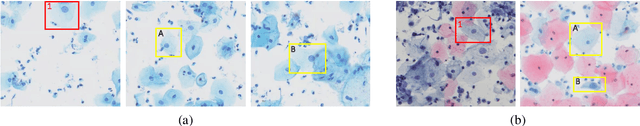
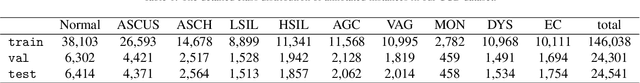
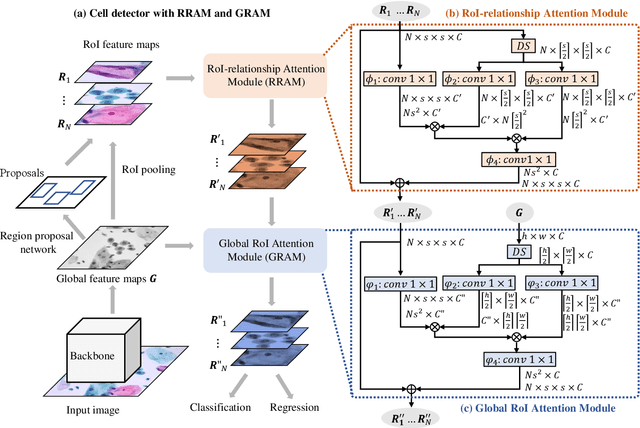
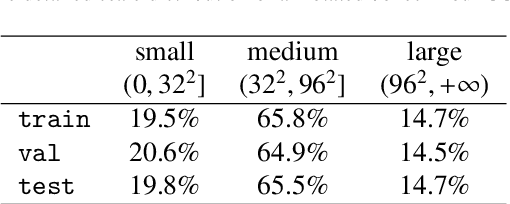
Abstract:Cervical abnormal cell detection is a challenging task as the morphological discrepancies between abnormal and normal cells are usually subtle. To determine whether a cervical cell is normal or abnormal, cytopathologists always take surrounding cells as references to identify its abnormality. To mimic these behaviors, we propose to explore contextual relationships to boost the performance of cervical abnormal cell detection. Specifically, both contextual relationships between cells and cell-to-global images are exploited to enhance features of each region of interest (RoI) proposals. Accordingly, two modules, dubbed as RoI-relationship attention module (RRAM) and global RoI attention module (GRAM), are developed and their combination strategies are also investigated. We establish a strong baseline by using Double-Head Faster R-CNN with feature pyramid network (FPN) and integrate our RRAM and GRAM into it to validate the effectiveness of the proposed modules. Experiments conducted on a large cervical cell detection dataset reveal that the introduction of RRAM and GRAM both achieves better average precision (AP) than the baseline methods. Moreover, when cascading RRAM and GRAM, our method outperforms the state-of-the-art (SOTA) methods. Furthermore, we also show the proposed feature enhancing scheme can facilitate both image-level and smear-level classification. The code and trained models are publicly available at https://github.com/CVIU-CSU/CR4CACD.
M2MRF: Many-to-Many Reassembly of Features for Tiny Lesion Segmentation in Fundus Images
Oct 30, 2021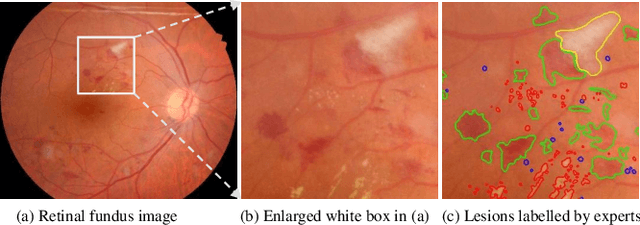
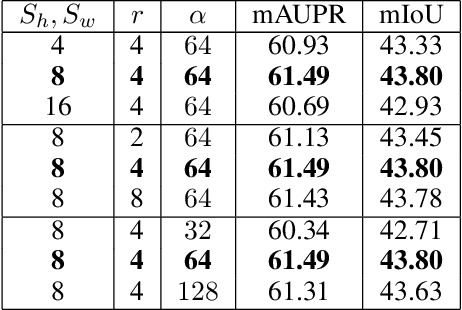
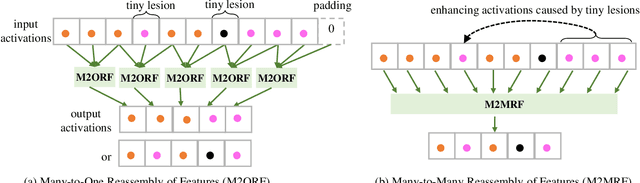
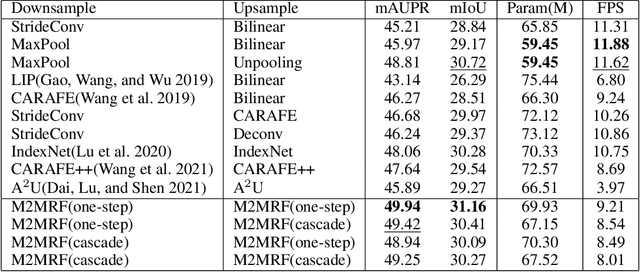
Abstract:Feature reassembly is an essential component in modern CNNs-based segmentation approaches, which includes feature downsampling and upsampling operators. Existing feature reassembly operators reassemble multiple features from a small predefined region into one for each target location independently. This may result in loss of spatial information, which could vanish activations of tiny lesions particularly when they cluster together. In this paper, we propose a many-to-many reassembly of features (M2MRF). It reassembles features in a dimension-reduced feature space and simultaneously aggregates multiple features inside a large predefined region into multiple target features. In this way, long range spatial dependencies are captured to maintain activations on tiny lesions, particularly when multiple lesions coexist. Experimental results on two lesion segmentation benchmarks, i.e. DDR and IDRiD, show that our M2MRF outperforms existing feature reassembly operators.
Dual-Branch Network with Dual-Sampling Modulated Dice Loss for Hard Exudate Segmentation from Colour Fundus Images
Dec 03, 2020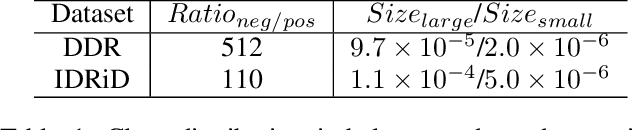
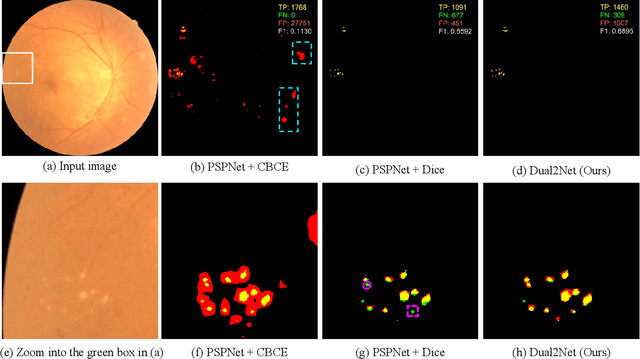
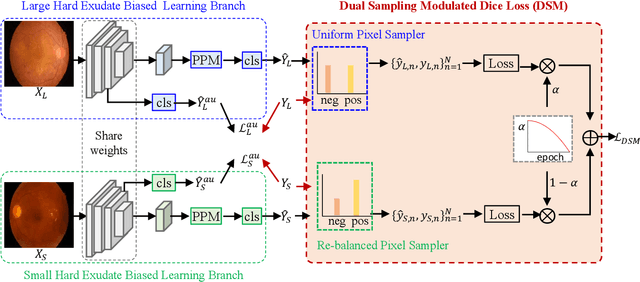
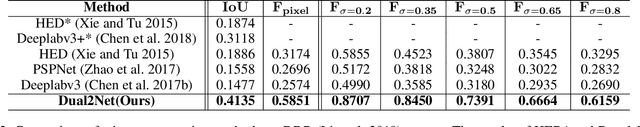
Abstract:Automated segmentation of hard exudates in colour fundus images is a challenge task due to issues of extreme class imbalance and enormous size variation. This paper aims to tackle these issues and proposes a dual-branch network with dual-sampling modulated Dice loss. It consists of two branches: large hard exudate biased learning branch and small hard exudate biased learning branch. Both of them are responsible for their own duty separately. Furthermore, we propose a dual-sampling modulated Dice loss for the training such that our proposed dual-branch network is able to segment hard exudates in different sizes. In detail, for the first branch, we use a uniform sampler to sample pixels from predicted segmentation mask for Dice loss calculation, which leads to this branch naturally be biased in favour of large hard exudates as Dice loss generates larger cost on misidentification of large hard exudates than small hard exudates. For the second branch, we use a re-balanced sampler to oversample hard exudate pixels and undersample background pixels for loss calculation. In this way, cost on misidentification of small hard exudates is enlarged, which enforces the parameters in the second branch fit small hard exudates well. Considering that large hard exudates are much easier to be correctly identified than small hard exudates, we propose an easy-to-difficult learning strategy by adaptively modulating the losses of two branches. We evaluate our proposed method on two public datasets and results demonstrate that ours achieves state-of-the-art performances.
A Novel Automation-Assisted Cervical Cancer Reading Method Based on Convolutional Neural Network
Dec 14, 2019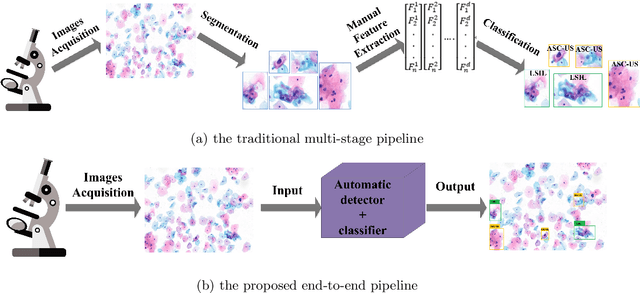
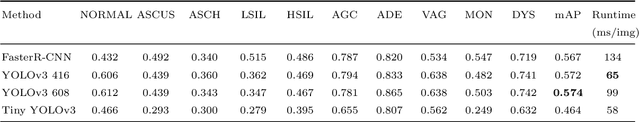
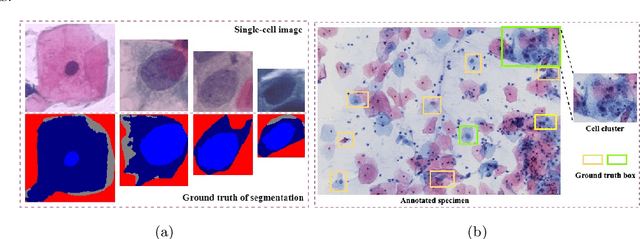
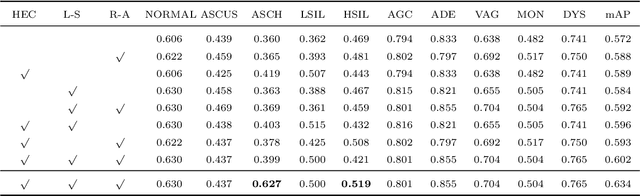
Abstract:While most previous automation-assisted reading methods can improve efficiency, their performance often relies on the success of accurate cell segmentation and hand-craft feature extraction. This paper presents an efficient and totally segmentation-free method for automated cervical cell screening that utilizes modern object detector to directly detect cervical cells or clumps, without the design of specific hand-crafted feature. Specifically, we use the state-of-the-art CNN-based object detection methods, YOLOv3, as our baseline model. In order to improve the classification performance of hard examples which are four highly similar categories, we cascade an additional task-specific classifier. We also investigate the presence of unreliable annotations and cope with them by smoothing the distribution of noisy labels. We comprehensively evaluate our methods on test set which is consisted of 1,014 annotated cervical cell images with size of 4000*3000 and complex cellular situation corresponding to 10 categories. Our model achieves 97.5% sensitivity (Sens) and 67.8% specificity (Spec) on cervical cell image-level screening. Moreover, we obtain a mean Average Precision (mAP) of 63.4% on cervical cell-level diagnosis, and improve the Average Precision (AP) of hard examples which are valuable but difficult to distinguish. Our automation-assisted cervical cell reading method not only achieves cervical cell image-level classification but also provides more detailed location and category information of abnormal cells. The results indicate feasible performance of our method, together with the efficiency and robustness, providing a new idea for future development of computer-assisted reading system in clinical cervical screening.
A Deep Gradient Boosting Network for Optic Disc and Cup Segmentation
Nov 05, 2019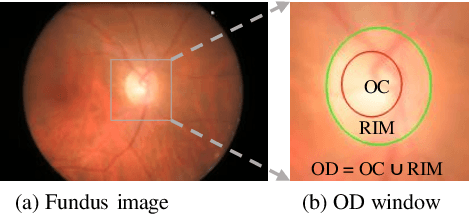
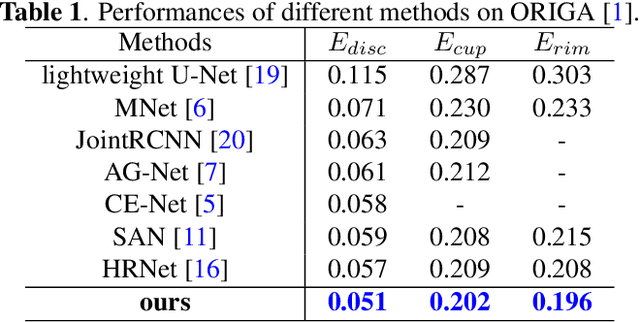
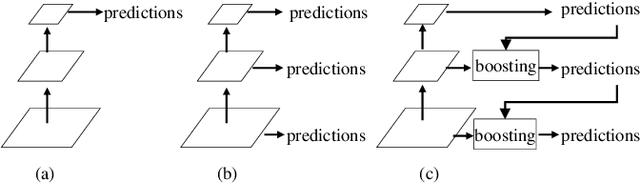
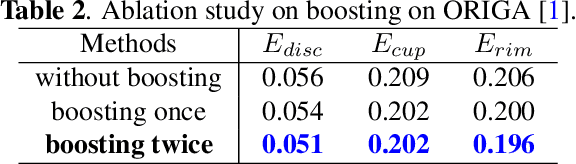
Abstract:Segmentation of optic disc (OD) and optic cup (OC) is critical in automated fundus image analysis system. Existing state-of-the-arts focus on designing deep neural networks with one or multiple dense prediction branches. Such kind of designs ignore connections among prediction branches and their learning capacity is limited. To build connections among prediction branches, this paper introduces gradient boosting framework to deep classification model and proposes a gradient boosting network called BoostNet. Specifically, deformable side-output unit and aggregation unit with deep supervisions are proposed to learn base functions and expansion coefficients in gradient boosting framework. By stacking aggregation units in a deep-to-shallow manner, models' performances are gradually boosted along deep to shallow stages. BoostNet achieves superior results to existing deep OD and OC segmentation networks on the public dataset ORIGA.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge