Yingyu Lin
Almost Linear Convergence under Minimal Score Assumptions: Quantized Transition Diffusion
May 28, 2025Abstract:Continuous diffusion models have demonstrated remarkable performance in data generation across various domains, yet their efficiency remains constrained by two critical limitations: (1) the local adjacency structure of the forward Markov process, which restricts long-range transitions in the data space, and (2) inherent biases introduced during the simulation of time-inhomogeneous reverse denoising processes. To address these challenges, we propose Quantized Transition Diffusion (QTD), a novel approach that integrates data quantization with discrete diffusion dynamics. Our method first transforms the continuous data distribution $p_*$ into a discrete one $q_*$ via histogram approximation and binary encoding, enabling efficient representation in a structured discrete latent space. We then design a continuous-time Markov chain (CTMC) with Hamming distance-based transitions as the forward process, which inherently supports long-range movements in the original data space. For reverse-time sampling, we introduce a \textit{truncated uniformization} technique to simulate the reverse CTMC, which can provably provide unbiased generation from $q_*$ under minimal score assumptions. Through a novel KL dynamic analysis of the reverse CTMC, we prove that QTD can generate samples with $O(d\ln^2(d/\epsilon))$ score evaluations in expectation to approximate the $d$--dimensional target distribution $p_*$ within an $\epsilon$ error tolerance. Our method not only establishes state-of-the-art inference efficiency but also advances the theoretical foundations of diffusion-based generative modeling by unifying discrete and continuous diffusion paradigms.
Purifying Approximate Differential Privacy with Randomized Post-processing
Mar 27, 2025Abstract:We propose a framework to convert $(\varepsilon, \delta)$-approximate Differential Privacy (DP) mechanisms into $(\varepsilon, 0)$-pure DP mechanisms, a process we call ``purification''. This algorithmic technique leverages randomized post-processing with calibrated noise to eliminate the $\delta$ parameter while preserving utility. By combining the tighter utility bounds and computational efficiency of approximate DP mechanisms with the stronger guarantees of pure DP, our approach achieves the best of both worlds. We illustrate the applicability of this framework in various settings, including Differentially Private Empirical Risk Minimization (DP-ERM), data-dependent DP mechanisms such as Propose-Test-Release (PTR), and query release tasks. To the best of our knowledge, this is the first work to provide a systematic method for transforming approximate DP into pure DP while maintaining competitive accuracy and computational efficiency.
A Skewness-Based Criterion for Addressing Heteroscedastic Noise in Causal Discovery
Oct 08, 2024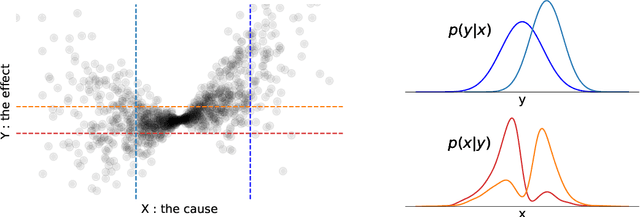

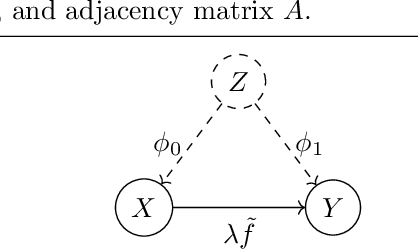

Abstract:Real-world data often violates the equal-variance assumption (homoscedasticity), making it essential to account for heteroscedastic noise in causal discovery. In this work, we explore heteroscedastic symmetric noise models (HSNMs), where the effect $Y$ is modeled as $Y = f(X) + \sigma(X)N$, with $X$ as the cause and $N$ as independent noise following a symmetric distribution. We introduce a novel criterion for identifying HSNMs based on the skewness of the score (i.e., the gradient of the log density) of the data distribution. This criterion establishes a computationally tractable measurement that is zero in the causal direction but nonzero in the anticausal direction, enabling the causal direction discovery. We extend this skewness-based criterion to the multivariate setting and propose SkewScore, an algorithm that handles heteroscedastic noise without requiring the extraction of exogenous noise. We also conduct a case study on the robustness of SkewScore in a bivariate model with a latent confounder, providing theoretical insights into its performance. Empirical studies further validate the effectiveness of the proposed method.
Tractable MCMC for Private Learning with Pure and Gaussian Differential Privacy
Oct 23, 2023Abstract:Posterior sampling, i.e., exponential mechanism to sample from the posterior distribution, provides $\varepsilon$-pure differential privacy (DP) guarantees and does not suffer from potentially unbounded privacy breach introduced by $(\varepsilon,\delta)$-approximate DP. In practice, however, one needs to apply approximate sampling methods such as Markov chain Monte Carlo (MCMC), thus re-introducing the unappealing $\delta$-approximation error into the privacy guarantees. To bridge this gap, we propose the Approximate SAample Perturbation (abbr. ASAP) algorithm which perturbs an MCMC sample with noise proportional to its Wasserstein-infinity ($W_\infty$) distance from a reference distribution that satisfies pure DP or pure Gaussian DP (i.e., $\delta=0$). We then leverage a Metropolis-Hastings algorithm to generate the sample and prove that the algorithm converges in W$_\infty$ distance. We show that by combining our new techniques with a careful localization step, we obtain the first nearly linear-time algorithm that achieves the optimal rates in the DP-ERM problem with strongly convex and smooth losses.
Leveraging Prior Knowledge for Protein-Protein Interaction Extraction with Memory Network
Jan 07, 2020
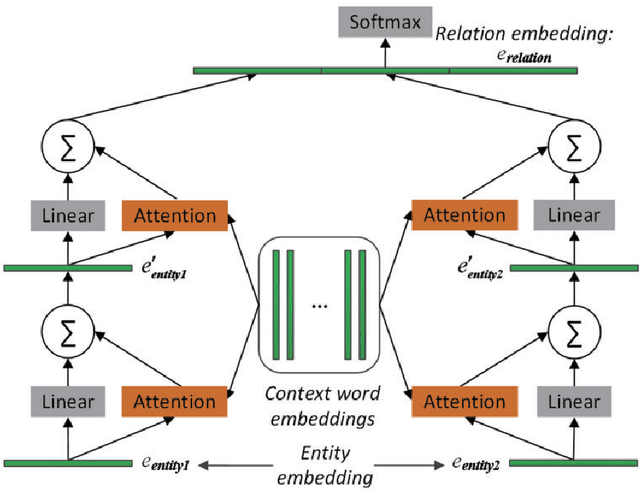
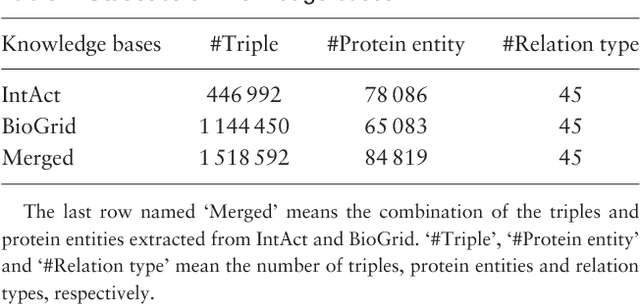
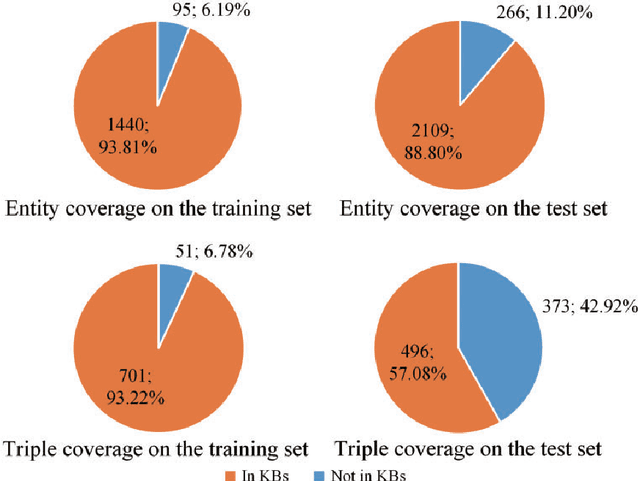
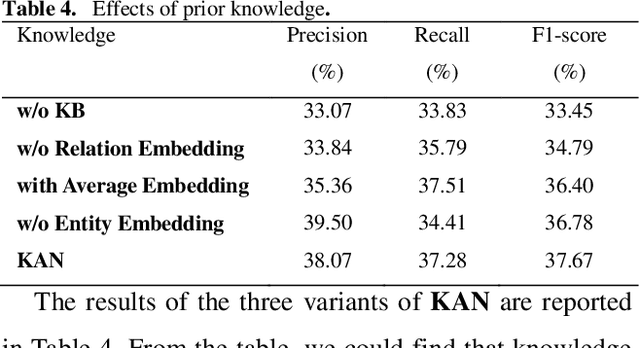
Abstract:Automatically extracting Protein-Protein Interactions (PPI) from biomedical literature provides additional support for precision medicine efforts. This paper proposes a novel memory network-based model (MNM) for PPI extraction, which leverages prior knowledge about protein-protein pairs with memory networks. The proposed MNM captures important context clues related to knowledge representations learned from knowledge bases. Both entity embeddings and relation embeddings of prior knowledge are effective in improving the PPI extraction model, leading to a new state-of-the-art performance on the BioCreative VI PPI dataset. The paper also shows that multiple computational layers over an external memory are superior to long short-term memory networks with the local memories.
* Published on Database-The Journal of Biological Databases and Curation, 11 pages, 5 figures
Knowledge-aware Attention Network for Protein-Protein Interaction Extraction
Jan 07, 2020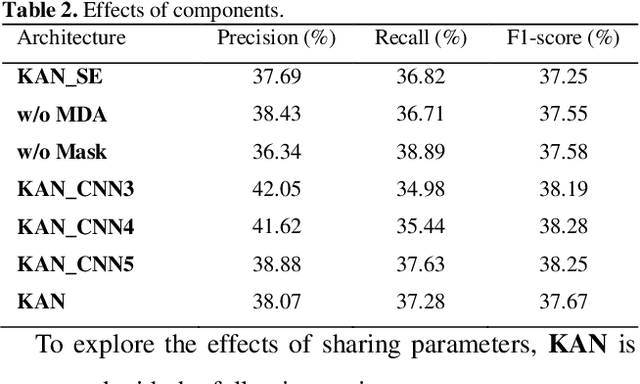
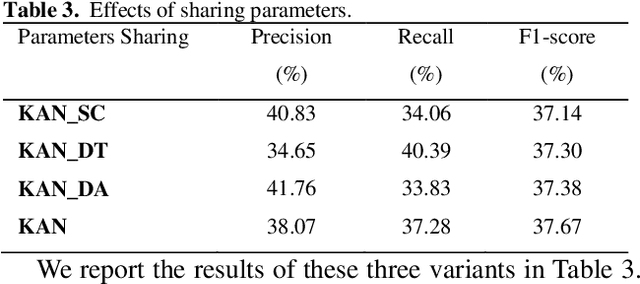
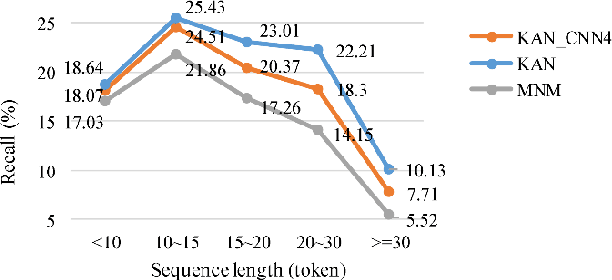
Abstract:Protein-protein interaction (PPI) extraction from published scientific literature provides additional support for precision medicine efforts. However, many of the current PPI extraction methods need extensive feature engineering and cannot make full use of the prior knowledge in knowledge bases (KB). KBs contain huge amounts of structured information about entities and relationships, therefore plays a pivotal role in PPI extraction. This paper proposes a knowledge-aware attention network (KAN) to fuse prior knowledge about protein-protein pairs and context information for PPI extraction. The proposed model first adopts a diagonal-disabled multi-head attention mechanism to encode context sequence along with knowledge representations learned from KB. Then a novel multi-dimensional attention mechanism is used to select the features that can best describe the encoded context. Experiment results on the BioCreative VI PPI dataset show that the proposed approach could acquire knowledge-aware dependencies between different words in a sequence and lead to a new state-of-the-art performance.
* Published on Journal of Biomedical Informatics, 14 pages, 5 figures
Chemical-induced Disease Relation Extraction with Dependency Information and Prior Knowledge
Jan 02, 2020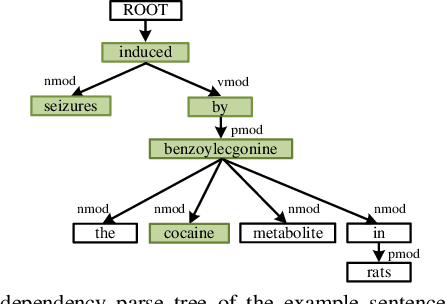
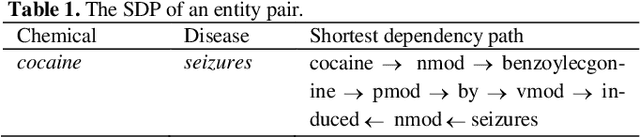
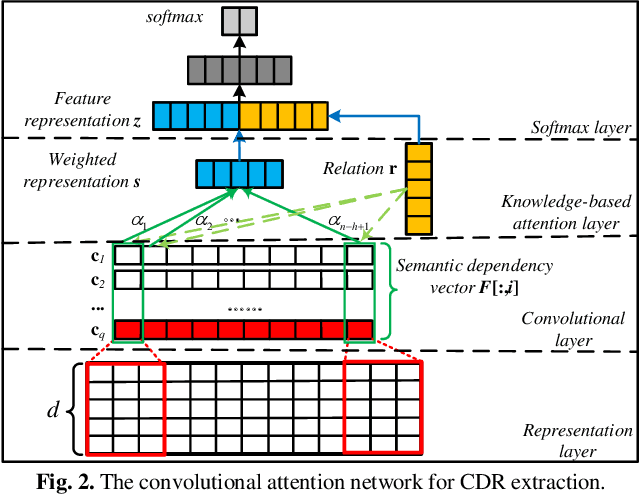
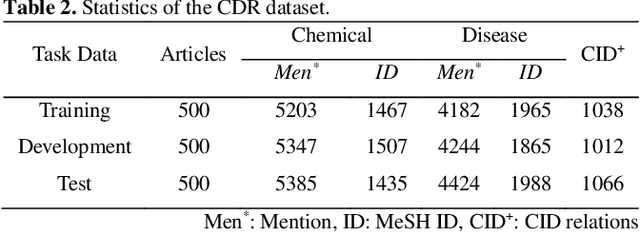
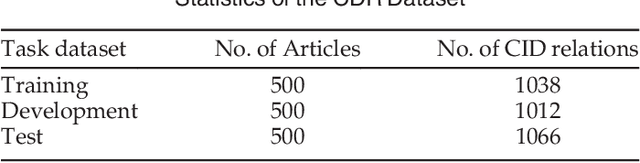
Abstract:Chemical-disease relation (CDR) extraction is significantly important to various areas of biomedical research and health care. Nowadays, many large-scale biomedical knowledge bases (KBs) containing triples about entity pairs and their relations have been built. KBs are important resources for biomedical relation extraction. However, previous research pays little attention to prior knowledge. In addition, the dependency tree contains important syntactic and semantic information, which helps to improve relation extraction. So how to effectively use it is also worth studying. In this paper, we propose a novel convolutional attention network (CAN) for CDR extraction. Firstly, we extract the shortest dependency path (SDP) between chemical and disease pairs in a sentence, which includes a sequence of words, dependency directions, and dependency relation tags. Then the convolution operations are performed on the SDP to produce deep semantic dependency features. After that, an attention mechanism is employed to learn the importance/weight of each semantic dependency vector related to knowledge representations learned from KBs. Finally, in order to combine dependency information and prior knowledge, the concatenation of weighted semantic dependency representations and knowledge representations is fed to the softmax layer for classification. Experiments on the BioCreative V CDR dataset show that our method achieves comparable performance with the state-of-the-art systems, and both dependency information and prior knowledge play important roles in CDR extraction task.
* Published on Journal of Biomedical Informatics, 13 pages
Combining Context and Knowledge Representations for Chemical-Disease Relation Extraction
Dec 23, 2019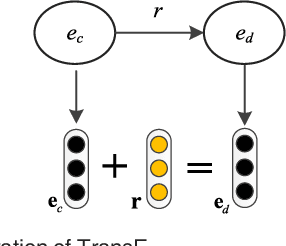
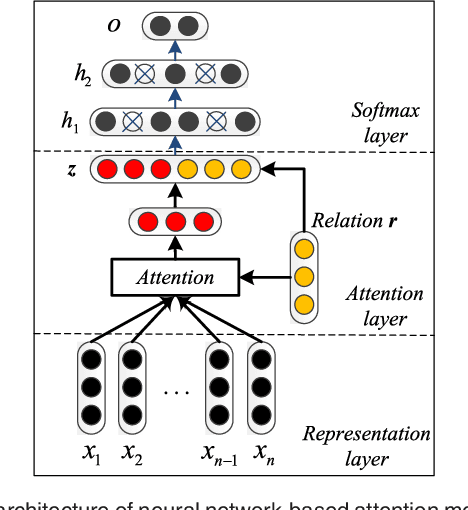
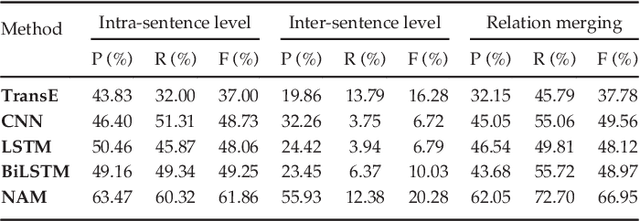
Abstract:Automatically extracting the relationships between chemicals and diseases is significantly important to various areas of biomedical research and health care. Biomedical experts have built many large-scale knowledge bases (KBs) to advance the development of biomedical research. KBs contain huge amounts of structured information about entities and relationships, therefore plays a pivotal role in chemical-disease relation (CDR) extraction. However, previous researches pay less attention to the prior knowledge existing in KBs. This paper proposes a neural network-based attention model (NAM) for CDR extraction, which makes full use of context information in documents and prior knowledge in KBs. For a pair of entities in a document, an attention mechanism is employed to select important context words with respect to the relation representations learned from KBs. Experiments on the BioCreative V CDR dataset show that combining context and knowledge representations through the attention mechanism, could significantly improve the CDR extraction performance while achieve comparable results with state-of-the-art systems.
* Published on IEEE/ACM Transactions on Computational Biology and Bioinformatics, 11 pages, 5 figures
Knowledge-guided Convolutional Networks for Chemical-Disease Relation Extraction
Dec 23, 2019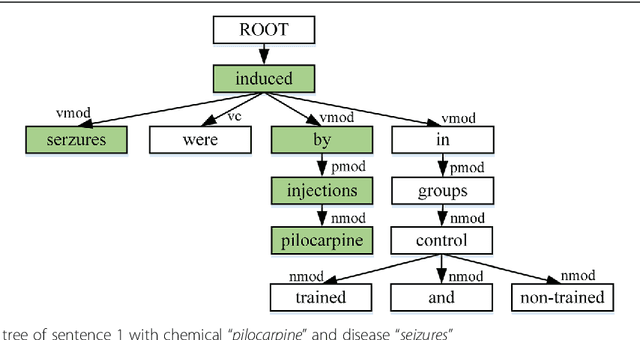
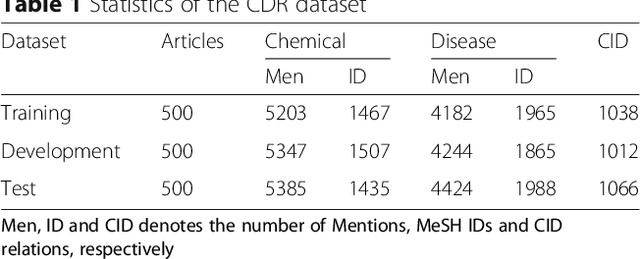
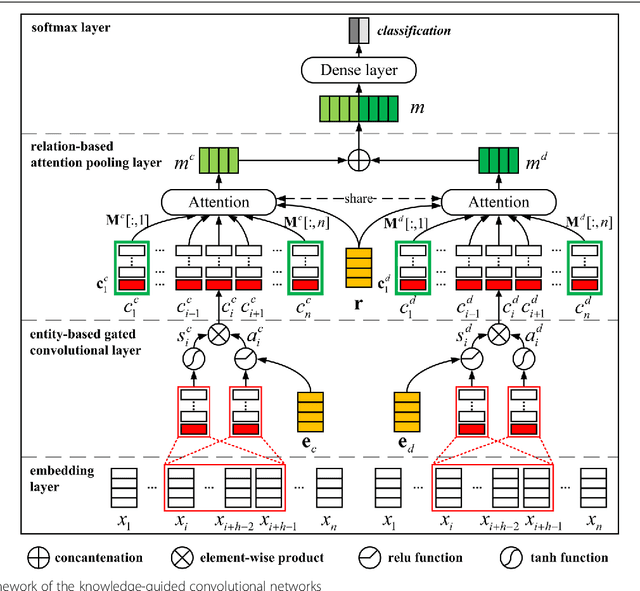
Abstract:Background: Automatic extraction of chemical-disease relations (CDR) from unstructured text is of essential importance for disease treatment and drug development. Meanwhile, biomedical experts have built many highly-structured knowledge bases (KBs), which contain prior knowledge about chemicals and diseases. Prior knowledge provides strong support for CDR extraction. How to make full use of it is worth studying. Results: This paper proposes a novel model called "Knowledge-guided Convolutional Networks (KCN)" to leverage prior knowledge for CDR extraction. The proposed model first learns knowledge representations including entity embeddings and relation embeddings from KBs. Then, entity embeddings are used to control the propagation of context features towards a chemical-disease pair with gated convolutions. After that, relation embeddings are employed to further capture the weighted context features by a shared attention pooling. Finally, the weighted context features containing additional knowledge information are used for CDR extraction. Experiments on the BioCreative V CDR dataset show that the proposed KCN achieves 71.28% F1-score, which outperforms most of the state-of-the-art systems. Conclusions: This paper proposes a novel CDR extraction model KCN to make full use of prior knowledge. Experimental results demonstrate that KCN could effectively integrate prior knowledge and contexts for the performance improvement.
* Published on BMC Bioinformatics, 16 pages, 5 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge