Yunlong Yang
Leveraging Prior Knowledge for Protein-Protein Interaction Extraction with Memory Network
Jan 07, 2020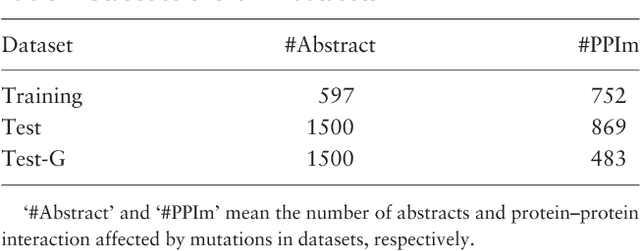
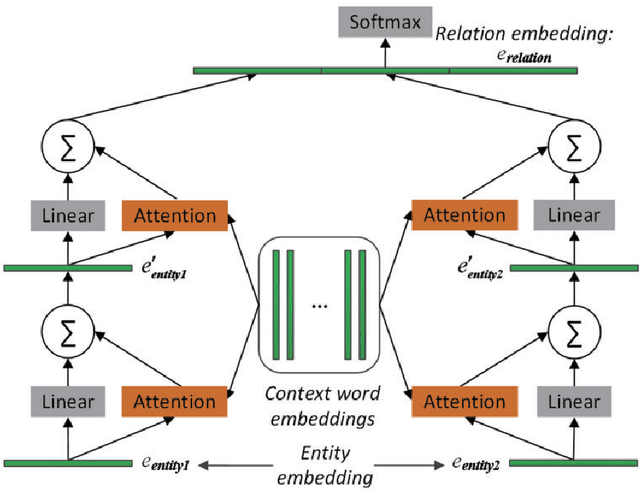
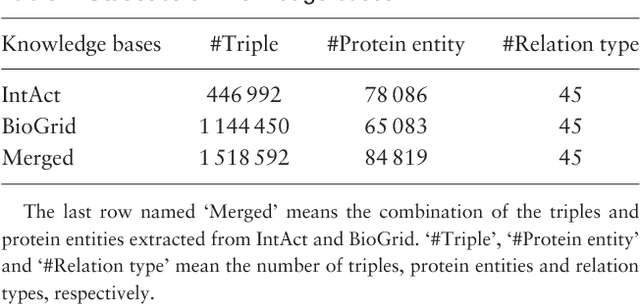
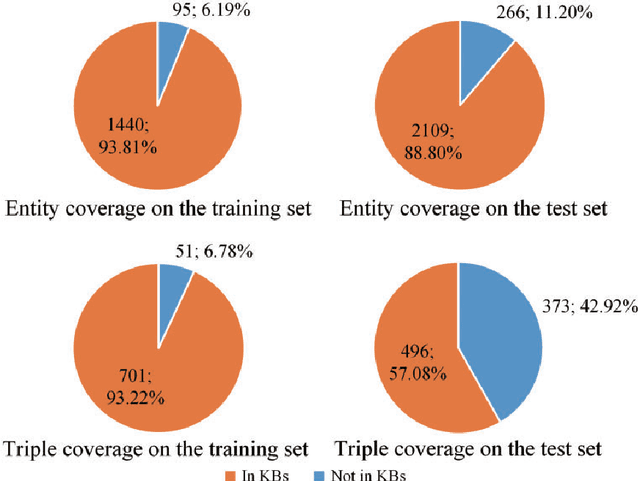
Abstract:Automatically extracting Protein-Protein Interactions (PPI) from biomedical literature provides additional support for precision medicine efforts. This paper proposes a novel memory network-based model (MNM) for PPI extraction, which leverages prior knowledge about protein-protein pairs with memory networks. The proposed MNM captures important context clues related to knowledge representations learned from knowledge bases. Both entity embeddings and relation embeddings of prior knowledge are effective in improving the PPI extraction model, leading to a new state-of-the-art performance on the BioCreative VI PPI dataset. The paper also shows that multiple computational layers over an external memory are superior to long short-term memory networks with the local memories.
* Published on Database-The Journal of Biological Databases and Curation, 11 pages, 5 figures
Chemical-induced Disease Relation Extraction with Dependency Information and Prior Knowledge
Jan 02, 2020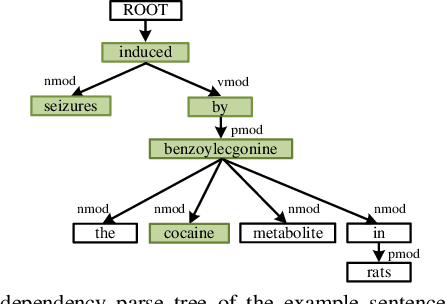
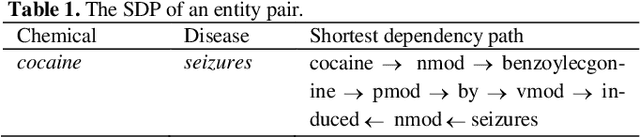
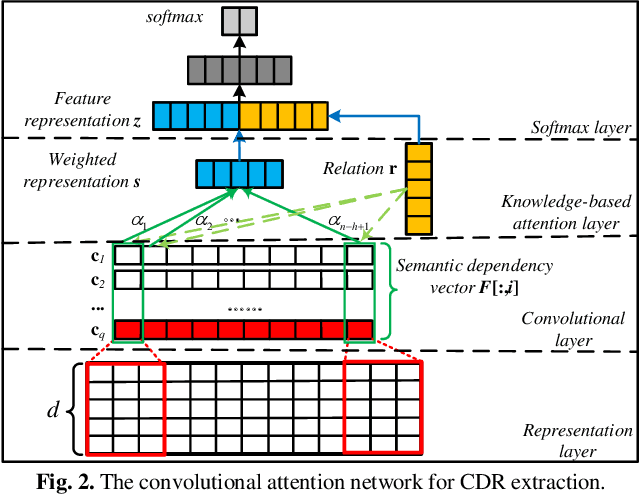
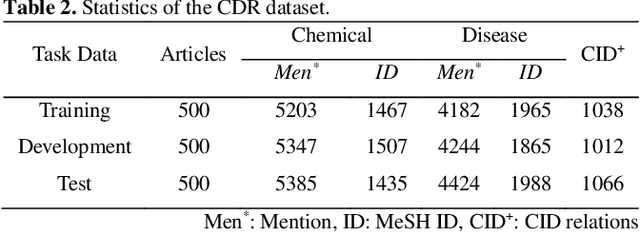
Abstract:Chemical-disease relation (CDR) extraction is significantly important to various areas of biomedical research and health care. Nowadays, many large-scale biomedical knowledge bases (KBs) containing triples about entity pairs and their relations have been built. KBs are important resources for biomedical relation extraction. However, previous research pays little attention to prior knowledge. In addition, the dependency tree contains important syntactic and semantic information, which helps to improve relation extraction. So how to effectively use it is also worth studying. In this paper, we propose a novel convolutional attention network (CAN) for CDR extraction. Firstly, we extract the shortest dependency path (SDP) between chemical and disease pairs in a sentence, which includes a sequence of words, dependency directions, and dependency relation tags. Then the convolution operations are performed on the SDP to produce deep semantic dependency features. After that, an attention mechanism is employed to learn the importance/weight of each semantic dependency vector related to knowledge representations learned from KBs. Finally, in order to combine dependency information and prior knowledge, the concatenation of weighted semantic dependency representations and knowledge representations is fed to the softmax layer for classification. Experiments on the BioCreative V CDR dataset show that our method achieves comparable performance with the state-of-the-art systems, and both dependency information and prior knowledge play important roles in CDR extraction task.
* Published on Journal of Biomedical Informatics, 13 pages
Combining Context and Knowledge Representations for Chemical-Disease Relation Extraction
Dec 23, 2019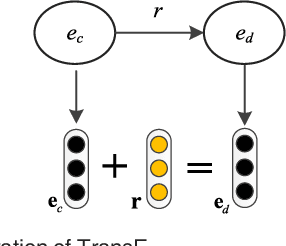
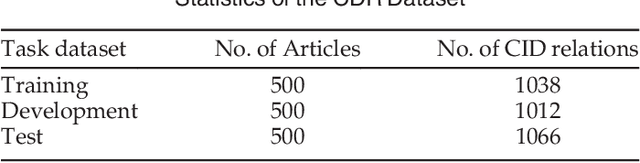
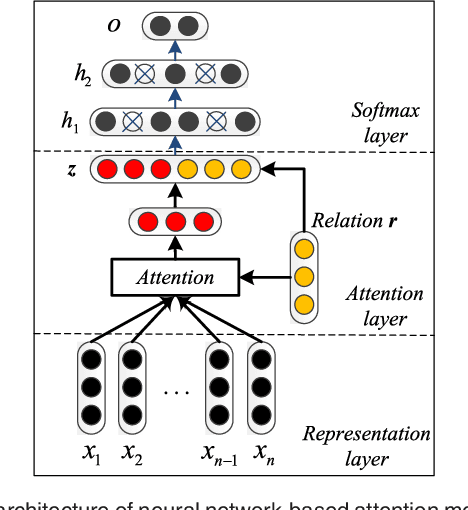
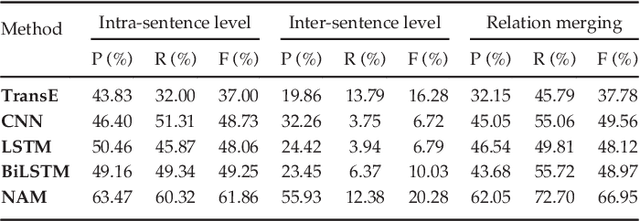
Abstract:Automatically extracting the relationships between chemicals and diseases is significantly important to various areas of biomedical research and health care. Biomedical experts have built many large-scale knowledge bases (KBs) to advance the development of biomedical research. KBs contain huge amounts of structured information about entities and relationships, therefore plays a pivotal role in chemical-disease relation (CDR) extraction. However, previous researches pay less attention to the prior knowledge existing in KBs. This paper proposes a neural network-based attention model (NAM) for CDR extraction, which makes full use of context information in documents and prior knowledge in KBs. For a pair of entities in a document, an attention mechanism is employed to select important context words with respect to the relation representations learned from KBs. Experiments on the BioCreative V CDR dataset show that combining context and knowledge representations through the attention mechanism, could significantly improve the CDR extraction performance while achieve comparable results with state-of-the-art systems.
* Published on IEEE/ACM Transactions on Computational Biology and Bioinformatics, 11 pages, 5 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge