Kun Ma
DriveLaW:Unifying Planning and Video Generation in a Latent Driving World
Dec 31, 2025Abstract:World models have become crucial for autonomous driving, as they learn how scenarios evolve over time to address the long-tail challenges of the real world. However, current approaches relegate world models to limited roles: they operate within ostensibly unified architectures that still keep world prediction and motion planning as decoupled processes. To bridge this gap, we propose DriveLaW, a novel paradigm that unifies video generation and motion planning. By directly injecting the latent representation from its video generator into the planner, DriveLaW ensures inherent consistency between high-fidelity future generation and reliable trajectory planning. Specifically, DriveLaW consists of two core components: DriveLaW-Video, our powerful world model that generates high-fidelity forecasting with expressive latent representations, and DriveLaW-Act, a diffusion planner that generates consistent and reliable trajectories from the latent of DriveLaW-Video, with both components optimized by a three-stage progressive training strategy. The power of our unified paradigm is demonstrated by new state-of-the-art results across both tasks. DriveLaW not only advances video prediction significantly, surpassing best-performing work by 33.3% in FID and 1.8% in FVD, but also achieves a new record on the NAVSIM planning benchmark.
The Meta Distribution of the SIR in Joint Communication and Sensing Networks
Apr 02, 2024Abstract:In this paper, we introduce a novel mathematical framework for assessing the performance of joint communication and sensing (JCAS) in wireless networks, employing stochastic geometry as an analytical tool. We focus on deriving the meta distribution of the signal-to-interference ratio (SIR) for JCAS networks. This approach enables a fine-grained quantification of individual user or radar performance intrinsic to these networks. Our work involves the modeling of JCAS networks and the derivation of mathematical expressions for the JCAS SIR meta distribution. Through simulations, we validate both our theoretical analysis and illustrate how the JCAS SIR meta distribution varies with the network deployment density.
Pattern-wise Transparent Sequential Recommendation
Feb 29, 2024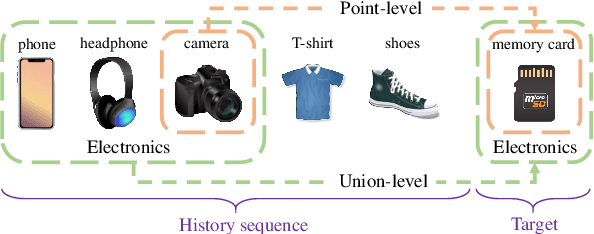
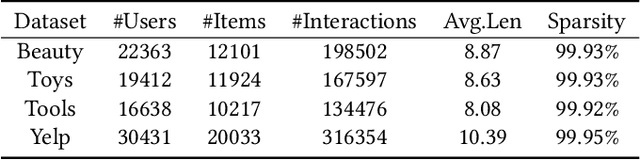
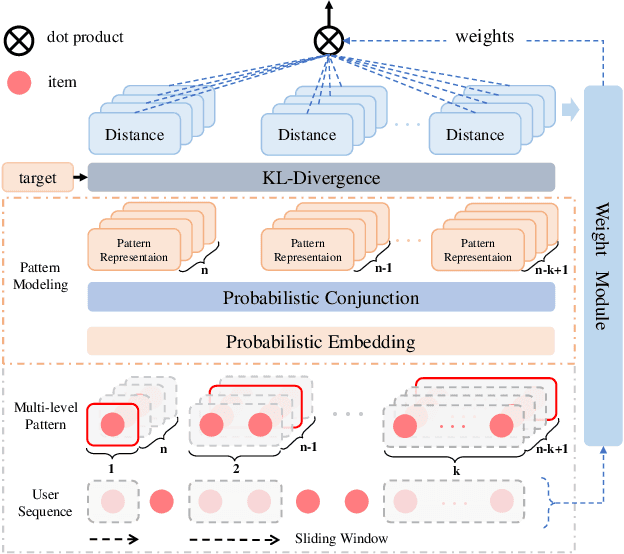
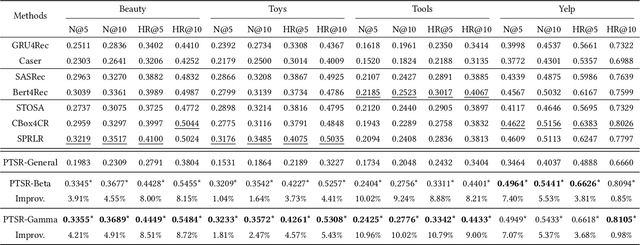
Abstract:A transparent decision-making process is essential for developing reliable and trustworthy recommender systems. For sequential recommendation, it means that the model can identify critical items asthe justifications for its recommendation results. However, achieving both model transparency and recommendation performance simultaneously is challenging, especially for models that take the entire sequence of items as input without screening. In this paper,we propose an interpretable framework (named PTSR) that enables a pattern-wise transparent decision-making process. It breaks the sequence of items into multi-level patterns that serve as atomic units for the entire recommendation process. The contribution of each pattern to the outcome is quantified in the probability space. With a carefully designed pattern weighting correction, the pattern contribution can be learned in the absence of ground-truth critical patterns. The final recommended items are those items that most critical patterns strongly endorse. Extensive experiments on four public datasets demonstrate remarkable recommendation performance, while case studies validate the model transparency. Our code is available at https://anonymous.4open.science/r/PTSR-2237.
Leveraging Prior Knowledge for Protein-Protein Interaction Extraction with Memory Network
Jan 07, 2020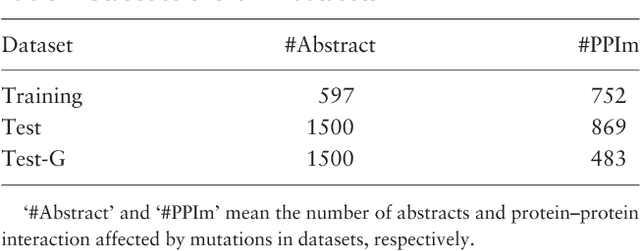
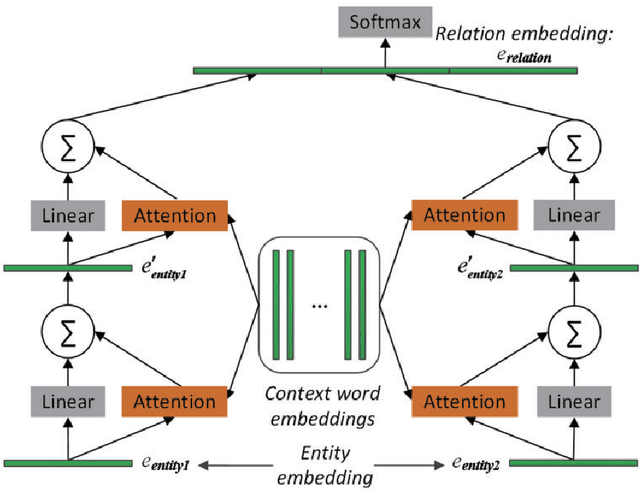
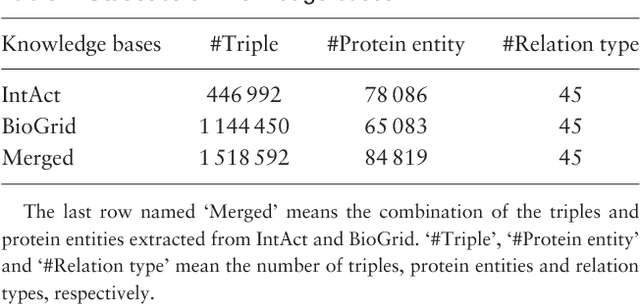
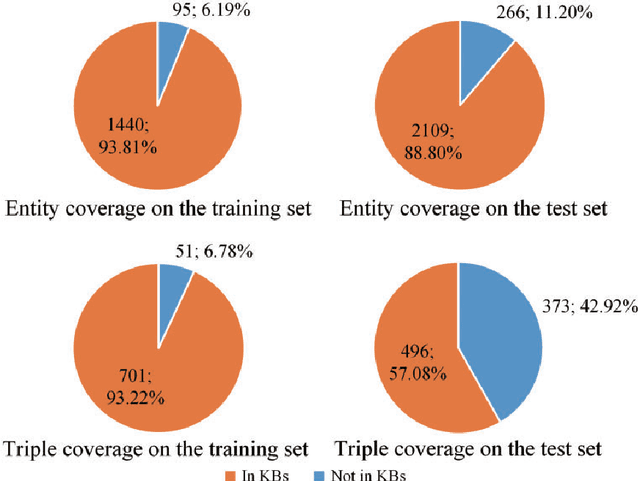
Abstract:Automatically extracting Protein-Protein Interactions (PPI) from biomedical literature provides additional support for precision medicine efforts. This paper proposes a novel memory network-based model (MNM) for PPI extraction, which leverages prior knowledge about protein-protein pairs with memory networks. The proposed MNM captures important context clues related to knowledge representations learned from knowledge bases. Both entity embeddings and relation embeddings of prior knowledge are effective in improving the PPI extraction model, leading to a new state-of-the-art performance on the BioCreative VI PPI dataset. The paper also shows that multiple computational layers over an external memory are superior to long short-term memory networks with the local memories.
* Published on Database-The Journal of Biological Databases and Curation, 11 pages, 5 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge