Vibhor Gupta
Jack
The Llama 4 Herd: Architecture, Training, Evaluation, and Deployment Notes
Jan 15, 2026Abstract:This document consolidates publicly reported technical details about Metas Llama 4 model family. It summarizes (i) released variants (Scout and Maverick) and the broader herd context including the previewed Behemoth teacher model, (ii) architectural characteristics beyond a high-level MoE description covering routed/shared-expert structure, early-fusion multimodality, and long-context design elements reported for Scout (iRoPE and length generalization strategies), (iii) training disclosures spanning pre-training, mid-training for long-context extension, and post-training methodology (lightweight SFT, online RL, and lightweight DPO) as described in release materials, (iv) developer-reported benchmark results for both base and instruction-tuned checkpoints, and (v) practical deployment constraints observed across major serving environments, including provider-specific context limits and quantization packaging. The manuscript also summarizes licensing obligations relevant to redistribution and derivative naming, and reviews publicly described safeguards and evaluation practices. The goal is to provide a compact technical reference for researchers and practitioners who need precise, source-backed facts about Llama 4.
The Llama 3 Herd of Models
Jul 31, 2024Abstract:Modern artificial intelligence (AI) systems are powered by foundation models. This paper presents a new set of foundation models, called Llama 3. It is a herd of language models that natively support multilinguality, coding, reasoning, and tool usage. Our largest model is a dense Transformer with 405B parameters and a context window of up to 128K tokens. This paper presents an extensive empirical evaluation of Llama 3. We find that Llama 3 delivers comparable quality to leading language models such as GPT-4 on a plethora of tasks. We publicly release Llama 3, including pre-trained and post-trained versions of the 405B parameter language model and our Llama Guard 3 model for input and output safety. The paper also presents the results of experiments in which we integrate image, video, and speech capabilities into Llama 3 via a compositional approach. We observe this approach performs competitively with the state-of-the-art on image, video, and speech recognition tasks. The resulting models are not yet being broadly released as they are still under development.
The Potential and Pitfalls of using a Large Language Model such as ChatGPT or GPT-4 as a Clinical Assistant
Jul 16, 2023
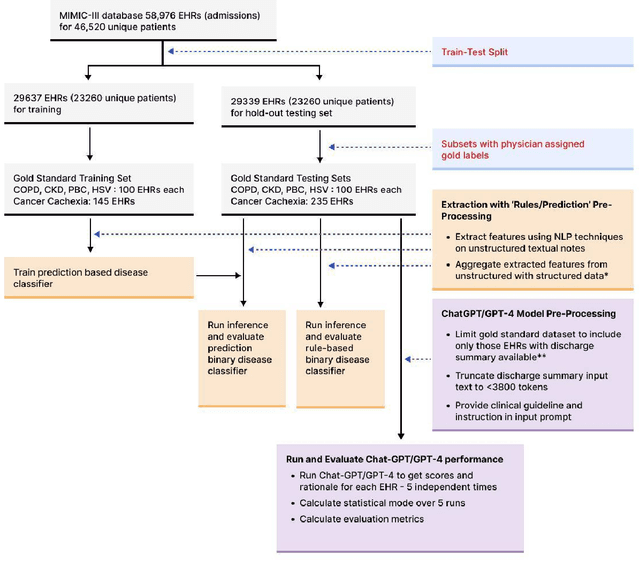
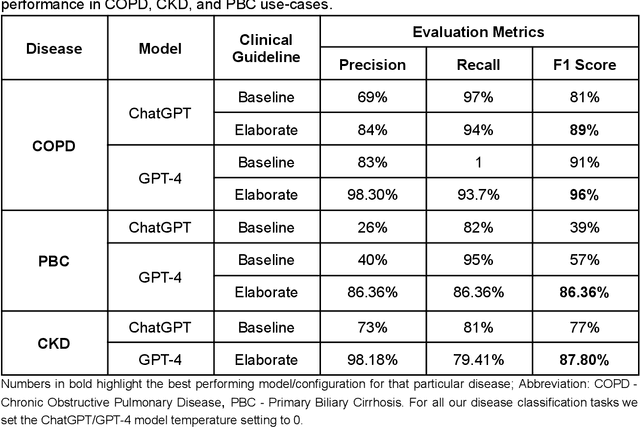
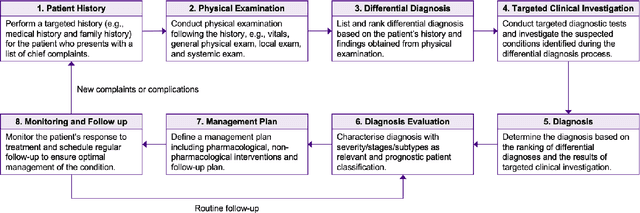
Abstract:Recent studies have demonstrated promising performance of ChatGPT and GPT-4 on several medical domain tasks. However, none have assessed its performance using a large-scale real-world electronic health record database, nor have evaluated its utility in providing clinical diagnostic assistance for patients across a full range of disease presentation. We performed two analyses using ChatGPT and GPT-4, one to identify patients with specific medical diagnoses using a real-world large electronic health record database and the other, in providing diagnostic assistance to healthcare workers in the prospective evaluation of hypothetical patients. Our results show that GPT-4 across disease classification tasks with chain of thought and few-shot prompting can achieve performance as high as 96% F1 scores. For patient assessment, GPT-4 can accurately diagnose three out of four times. However, there were mentions of factually incorrect statements, overlooking crucial medical findings, recommendations for unnecessary investigations and overtreatment. These issues coupled with privacy concerns, make these models currently inadequate for real world clinical use. However, limited data and time needed for prompt engineering in comparison to configuration of conventional machine learning workflows highlight their potential for scalability across healthcare applications.
Medical Scientific Table-to-Text Generation with Human-in-the-Loop under the Data Sparsity Constraint
May 24, 2022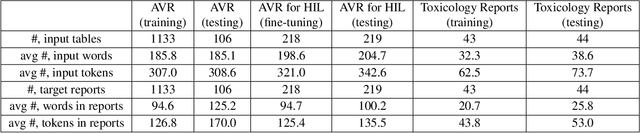
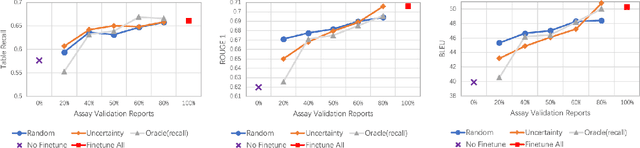
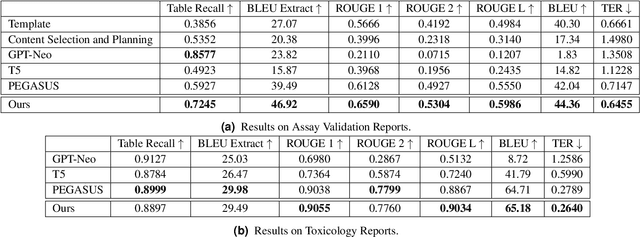
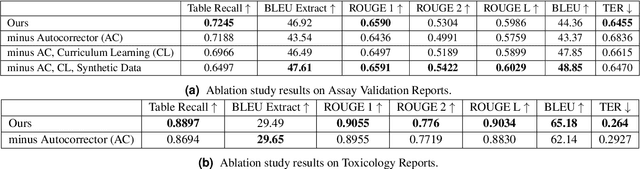
Abstract:Structured (tabular) data in the preclinical and clinical domains contains valuable information about individuals and an efficient table-to-text summarization system can drastically reduce manual efforts to condense this data into reports. However, in practice, the problem is heavily impeded by the data paucity, data sparsity and inability of the state-of-the-art natural language generation models (including T5, PEGASUS and GPT-Neo) to produce accurate and reliable outputs. In this paper, we propose a novel table-to-text approach and tackle these problems with a novel two-step architecture which is enhanced by auto-correction, copy mechanism and synthetic data augmentation. The study shows that the proposed approach selects salient biomedical entities and values from structured data with improved precision (up to 0.13 absolute increase) of copying the tabular values to generate coherent and accurate text for assay validation reports and toxicology reports. Moreover, we also demonstrate a light-weight adaptation of the proposed system to new datasets by fine-tuning with as little as 40\% training examples. The outputs of our model are validated by human experts in the Human-in-the-Loop scenario.
A Scalable Workflow to Build Machine Learning Classifiers with Clinician-in-the-Loop to Identify Patients in Specific Diseases
May 18, 2022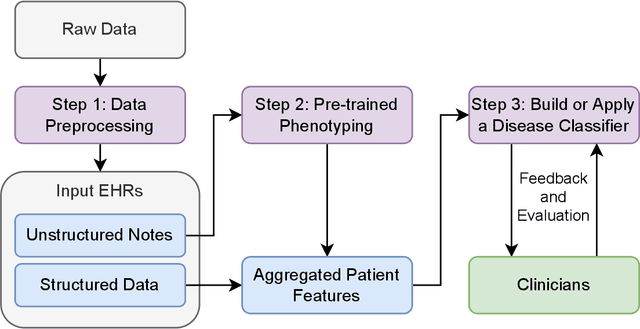

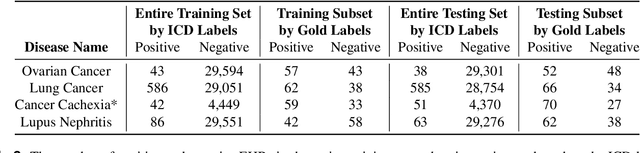
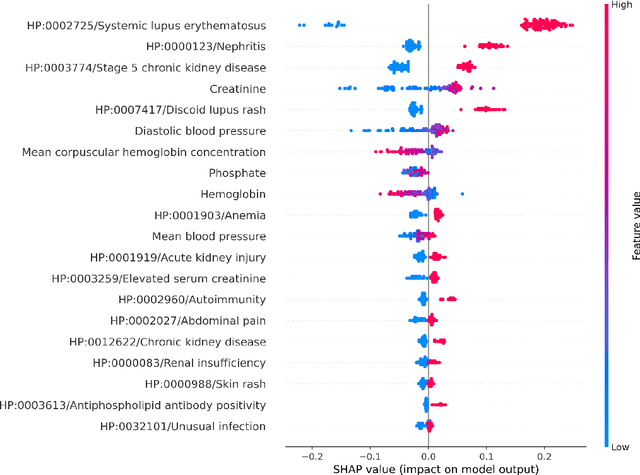
Abstract:Clinicians may rely on medical coding systems such as International Classification of Diseases (ICD) to identify patients with diseases from Electronic Health Records (EHRs). However, due to the lack of detail and specificity as well as a probability of miscoding, recent studies suggest the ICD codes often cannot characterise patients accurately for specific diseases in real clinical practice, and as a result, using them to find patients for studies or trials can result in high failure rates and missing out on uncoded patients. Manual inspection of all patients at scale is not feasible as it is highly costly and slow. This paper proposes a scalable workflow which leverages both structured data and unstructured textual notes from EHRs with techniques including NLP, AutoML and Clinician-in-the-Loop mechanism to build machine learning classifiers to identify patients at scale with given diseases, especially those who might currently be miscoded or missed by ICD codes. Case studies in the MIMIC-III dataset were conducted where the proposed workflow demonstrates a higher classification performance in terms of F1 scores compared to simply using ICD codes on gold testing subset to identify patients with Ovarian Cancer (0.901 vs 0.814), Lung Cancer (0.859 vs 0.828), Cancer Cachexia (0.862 vs 0.650), and Lupus Nephritis (0.959 vs 0.855). Also, the proposed workflow that leverages unstructured notes consistently outperforms the baseline that uses structured data only with an increase of F1 (Ovarian Cancer 0.901 vs 0.719, Lung Cancer 0.859 vs 0.787, Cancer Cachexia 0.862 vs 0.838 and Lupus Nephritis 0.959 vs 0.785). Experiments on the large testing set also demonstrate the proposed workflow can find more patients who are miscoded or missed by ICD codes. Moreover, interpretability studies are also conducted to clinically validate the top impact features of the classifiers.
Unsupervised Numerical Reasoning to Extract Phenotypes from Clinical Text by Leveraging External Knowledge
Apr 19, 2022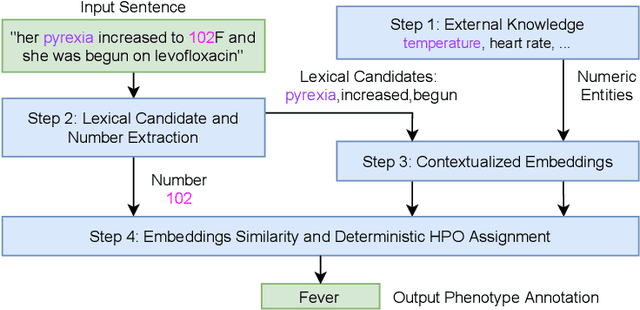
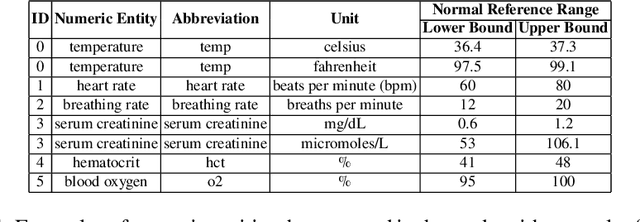
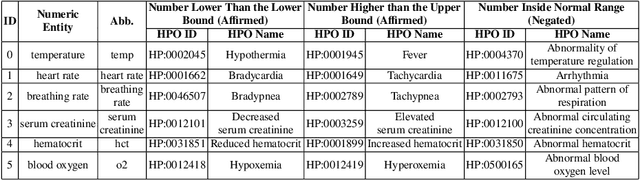
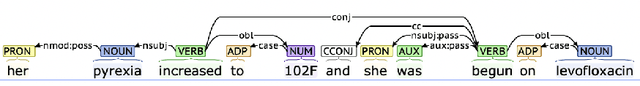
Abstract:Extracting phenotypes from clinical text has been shown to be useful for a variety of clinical use cases such as identifying patients with rare diseases. However, reasoning with numerical values remains challenging for phenotyping in clinical text, for example, temperature 102F representing Fever. Current state-of-the-art phenotyping models are able to detect general phenotypes, but perform poorly when they detect phenotypes requiring numerical reasoning. We present a novel unsupervised methodology leveraging external knowledge and contextualized word embeddings from ClinicalBERT for numerical reasoning in a variety of phenotypic contexts. Comparing against unsupervised benchmarks, it shows a substantial performance improvement with absolute gains on generalized Recall and F1 scores up to 79% and 71%, respectively. In the supervised setting, it also surpasses the performance of alternative approaches with absolute gains on generalized Recall and F1 scores up to 70% and 44%, respectively.
Continual Learning for Multivariate Time Series Tasks with Variable Input Dimensions
Mar 14, 2022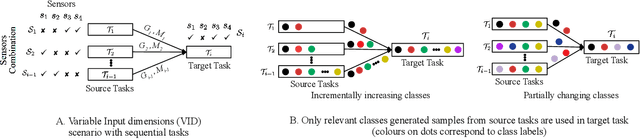
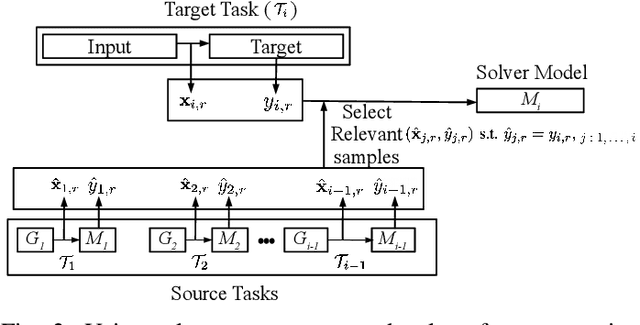
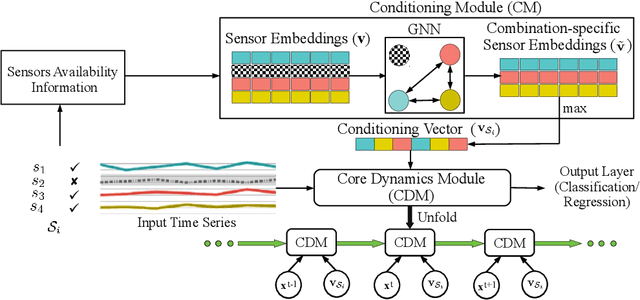
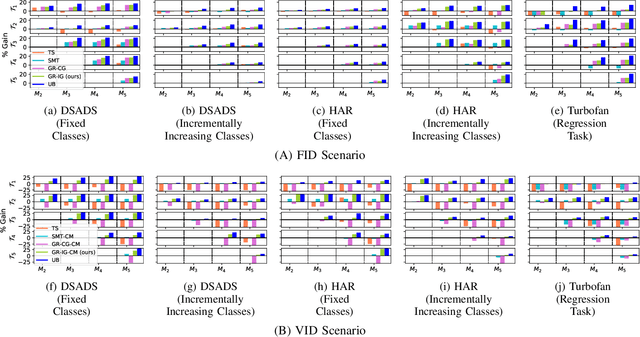
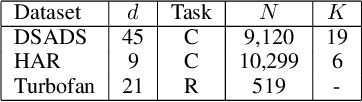
Abstract:We consider a sequence of related multivariate time series learning tasks, such as predicting failures for different instances of a machine from time series of multi-sensor data, or activity recognition tasks over different individuals from multiple wearable sensors. We focus on two under-explored practical challenges arising in such settings: (i) Each task may have a different subset of sensors, i.e., providing different partial observations of the underlying 'system'. This restriction can be due to different manufacturers in the former case, and people wearing more or less measurement devices in the latter (ii) We are not allowed to store or re-access data from a task once it has been observed at the task level. This may be due to privacy considerations in the case of people, or legal restrictions placed by machine owners. Nevertheless, we would like to (a) improve performance on subsequent tasks using experience from completed tasks as well as (b) continue to perform better on past tasks, e.g., update the model and improve predictions on even the first machine after learning from subsequently observed ones. We note that existing continual learning methods do not take into account variability in input dimensions arising due to different subsets of sensors being available across tasks, and struggle to adapt to such variable input dimensions (VID) tasks. In this work, we address this shortcoming of existing methods. To this end, we learn task-specific generative models and classifiers, and use these to augment data for target tasks. Since the input dimensions across tasks vary, we propose a novel conditioning module based on graph neural networks to aid a standard recurrent neural network. We evaluate the efficacy of the proposed approach on three publicly available datasets corresponding to two activity recognition tasks (classification) and one prognostics task (regression).
Self-Supervised Detection of Contextual Synonyms in a Multi-Class Setting: Phenotype Annotation Use Case
Sep 04, 2021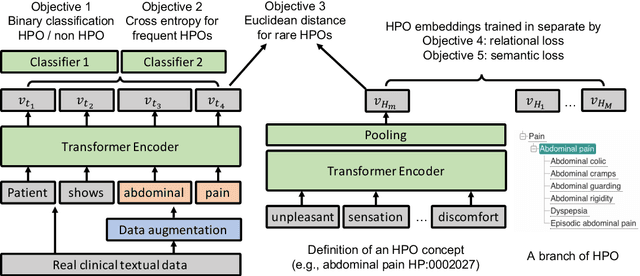
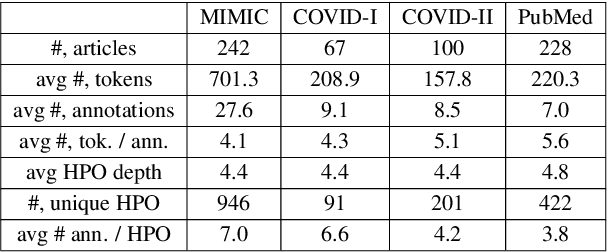
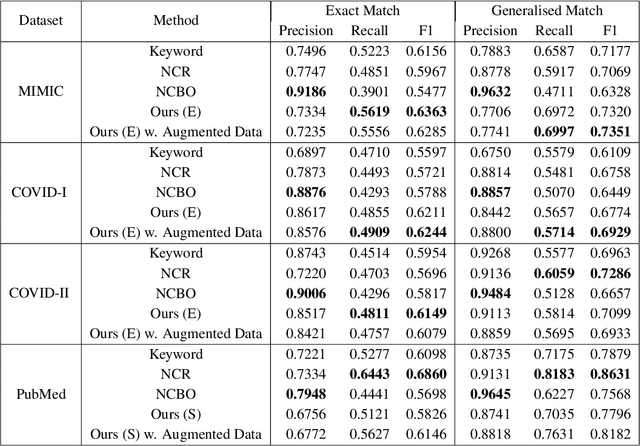
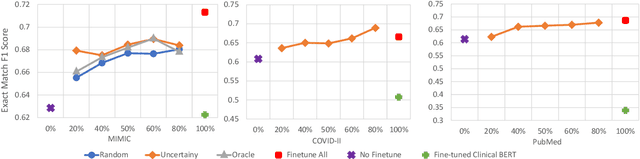
Abstract:Contextualised word embeddings is a powerful tool to detect contextual synonyms. However, most of the current state-of-the-art (SOTA) deep learning concept extraction methods remain supervised and underexploit the potential of the context. In this paper, we propose a self-supervised pre-training approach which is able to detect contextual synonyms of concepts being training on the data created by shallow matching. We apply our methodology in the sparse multi-class setting (over 15,000 concepts) to extract phenotype information from electronic health records. We further investigate data augmentation techniques to address the problem of the class sparsity. Our approach achieves a new SOTA for the unsupervised phenotype concept annotation on clinical text on F1 and Recall outperforming the previous SOTA with a gain of up to 4.5 and 4.0 absolute points, respectively. After fine-tuning with as little as 20\% of the labelled data, we also outperform BioBERT and ClinicalBERT. The extrinsic evaluation on three ICU benchmarks also shows the benefit of using the phenotypes annotated by our model as features.
Clinical Utility of the Automatic Phenotype Annotation in Unstructured Clinical Notes: ICU Use Cases
Jul 24, 2021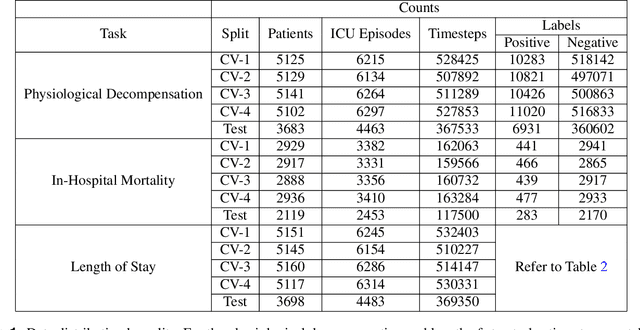
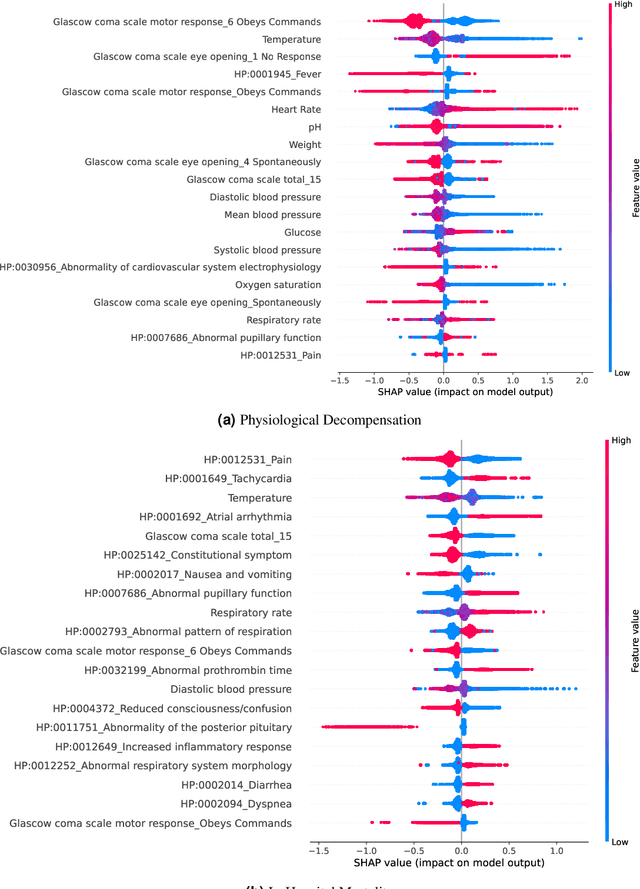
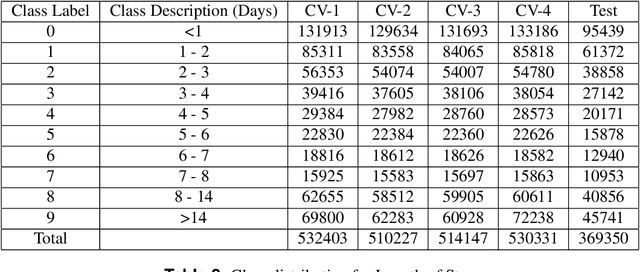
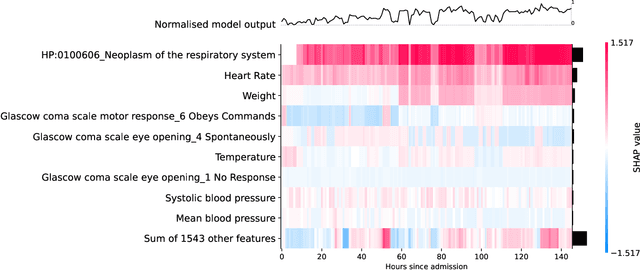
Abstract:Clinical notes contain information not present elsewhere, including drug response and symptoms, all of which are highly important when predicting key outcomes in acute care patients. We propose the automatic annotation of phenotypes from clinical notes as a method to capture essential information to predict outcomes in the Intensive Care Unit (ICU). This information is complementary to typically used vital signs and laboratory test results. We demonstrate and validate our approach conducting experiments on the prediction of in-hospital mortality, physiological decompensation and length of stay in the ICU setting for over 24,000 patients. The prediction models incorporating phenotypic information consistently outperform the baseline models leveraging only vital signs and laboratory test results. Moreover, we conduct a thorough interpretability study, showing that phenotypes provide valuable insights at the patient and cohort levels. Our approach illustrates the viability of using phenotypes to determine outcomes in the ICU.
Handling Variable-Dimensional Time Series with Graph Neural Networks
Jul 07, 2020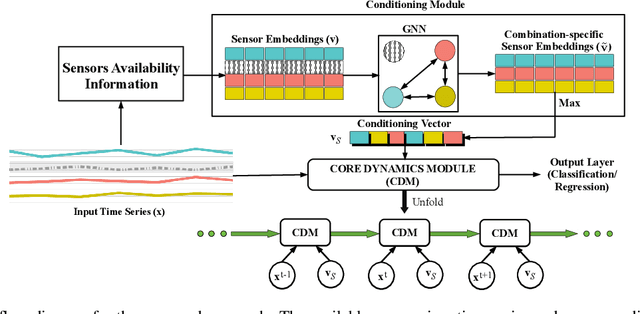
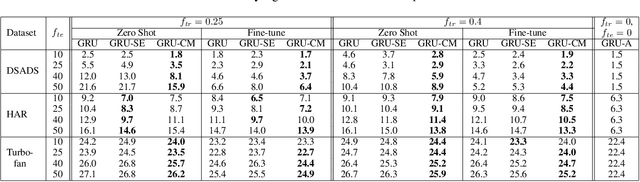
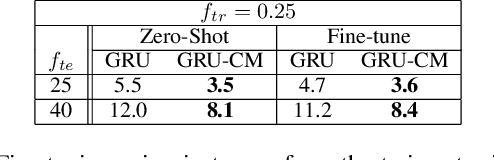
Abstract:Several applications of Internet of Things (IoT) technology involve capturing data from multiple sensors resulting in multi-sensor time series. Existing neural networks based approaches for such multi-sensor or multivariate time series modeling assume fixed input dimension or number of sensors. Such approaches can struggle in the practical setting where different instances of the same device or equipment such as mobiles, wearables, engines, etc. come with different combinations of installed sensors. We consider training neural network models from such multi-sensor time series, where the time series have varying input dimensionality owing to availability or installation of a different subset of sensors at each source of time series. We propose a novel neural network architecture suitable for zero-shot transfer learning allowing robust inference for multivariate time series with previously unseen combination of available dimensions or sensors at test time. Such a combinatorial generalization is achieved by conditioning the layers of a core neural network-based time series model with a "conditioning vector" that carries information of the available combination of sensors for each time series. This conditioning vector is obtained by summarizing the set of learned "sensor embedding vectors" corresponding to the available sensors in a time series via a graph neural network. We evaluate the proposed approach on publicly available activity recognition and equipment prognostics datasets, and show that the proposed approach allows for better generalization in comparison to a deep gated recurrent neural network baseline.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge