Luis Bolanos
Gaussian Shadow Casting for Neural Characters
Jan 11, 2024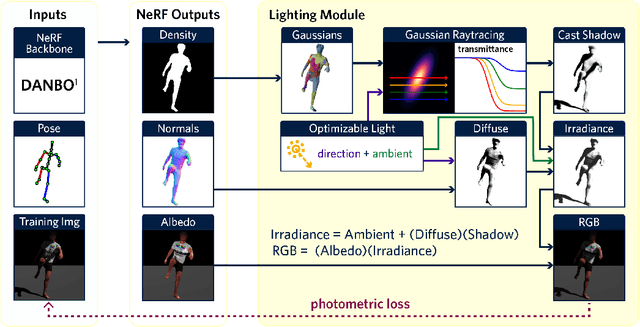


Abstract:Neural character models can now reconstruct detailed geometry and texture from video, but they lack explicit shadows and shading, leading to artifacts when generating novel views and poses or during relighting. It is particularly difficult to include shadows as they are a global effect and the required casting of secondary rays is costly. We propose a new shadow model using a Gaussian density proxy that replaces sampling with a simple analytic formula. It supports dynamic motion and is tailored for shadow computation, thereby avoiding the affine projection approximation and sorting required by the closely related Gaussian splatting. Combined with a deferred neural rendering model, our Gaussian shadows enable Lambertian shading and shadow casting with minimal overhead. We demonstrate improved reconstructions, with better separation of albedo, shading, and shadows in challenging outdoor scenes with direct sun light and hard shadows. Our method is able to optimize the light direction without any input from the user. As a result, novel poses have fewer shadow artifacts and relighting in novel scenes is more realistic compared to the state-of-the-art methods, providing new ways to pose neural characters in novel environments, increasing their applicability.
A Scalable Workflow to Build Machine Learning Classifiers with Clinician-in-the-Loop to Identify Patients in Specific Diseases
May 18, 2022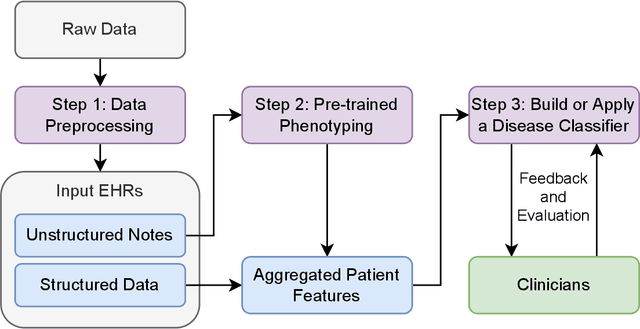

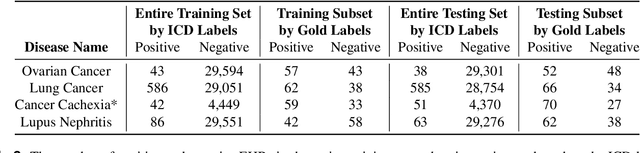
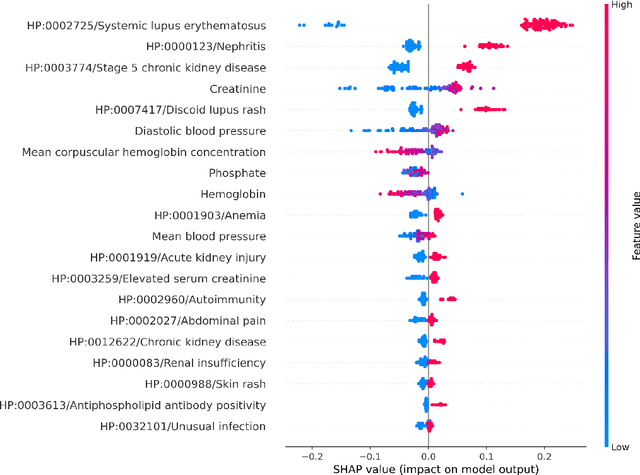
Abstract:Clinicians may rely on medical coding systems such as International Classification of Diseases (ICD) to identify patients with diseases from Electronic Health Records (EHRs). However, due to the lack of detail and specificity as well as a probability of miscoding, recent studies suggest the ICD codes often cannot characterise patients accurately for specific diseases in real clinical practice, and as a result, using them to find patients for studies or trials can result in high failure rates and missing out on uncoded patients. Manual inspection of all patients at scale is not feasible as it is highly costly and slow. This paper proposes a scalable workflow which leverages both structured data and unstructured textual notes from EHRs with techniques including NLP, AutoML and Clinician-in-the-Loop mechanism to build machine learning classifiers to identify patients at scale with given diseases, especially those who might currently be miscoded or missed by ICD codes. Case studies in the MIMIC-III dataset were conducted where the proposed workflow demonstrates a higher classification performance in terms of F1 scores compared to simply using ICD codes on gold testing subset to identify patients with Ovarian Cancer (0.901 vs 0.814), Lung Cancer (0.859 vs 0.828), Cancer Cachexia (0.862 vs 0.650), and Lupus Nephritis (0.959 vs 0.855). Also, the proposed workflow that leverages unstructured notes consistently outperforms the baseline that uses structured data only with an increase of F1 (Ovarian Cancer 0.901 vs 0.719, Lung Cancer 0.859 vs 0.787, Cancer Cachexia 0.862 vs 0.838 and Lupus Nephritis 0.959 vs 0.785). Experiments on the large testing set also demonstrate the proposed workflow can find more patients who are miscoded or missed by ICD codes. Moreover, interpretability studies are also conducted to clinically validate the top impact features of the classifiers.
Self-Supervised Detection of Contextual Synonyms in a Multi-Class Setting: Phenotype Annotation Use Case
Sep 04, 2021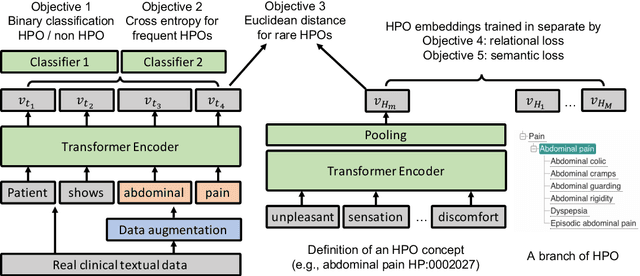
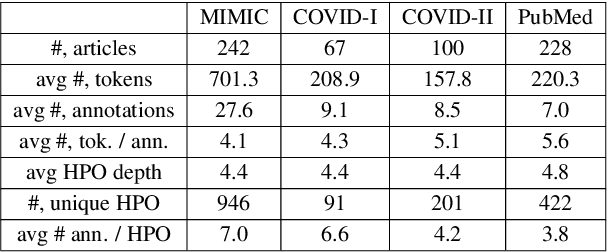
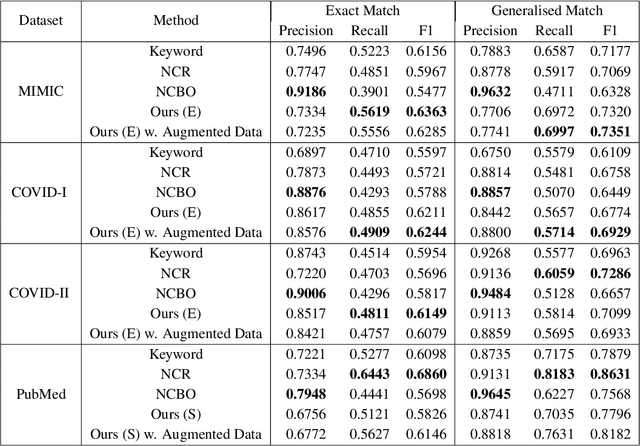
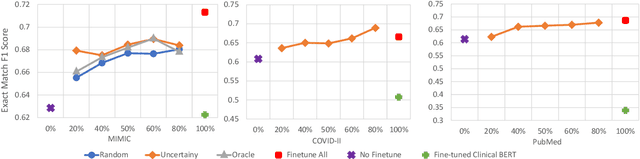
Abstract:Contextualised word embeddings is a powerful tool to detect contextual synonyms. However, most of the current state-of-the-art (SOTA) deep learning concept extraction methods remain supervised and underexploit the potential of the context. In this paper, we propose a self-supervised pre-training approach which is able to detect contextual synonyms of concepts being training on the data created by shallow matching. We apply our methodology in the sparse multi-class setting (over 15,000 concepts) to extract phenotype information from electronic health records. We further investigate data augmentation techniques to address the problem of the class sparsity. Our approach achieves a new SOTA for the unsupervised phenotype concept annotation on clinical text on F1 and Recall outperforming the previous SOTA with a gain of up to 4.5 and 4.0 absolute points, respectively. After fine-tuning with as little as 20\% of the labelled data, we also outperform BioBERT and ClinicalBERT. The extrinsic evaluation on three ICU benchmarks also shows the benefit of using the phenotypes annotated by our model as features.
Clinical Utility of the Automatic Phenotype Annotation in Unstructured Clinical Notes: ICU Use Cases
Jul 24, 2021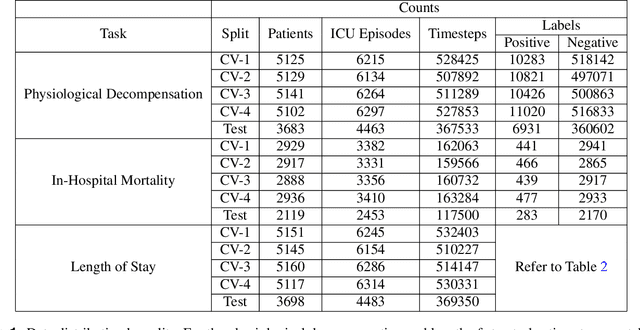
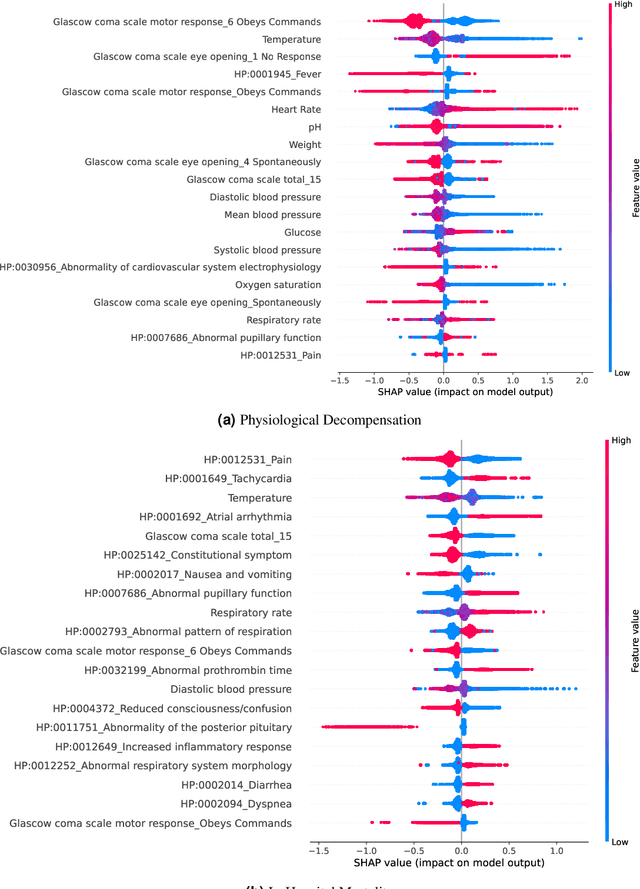
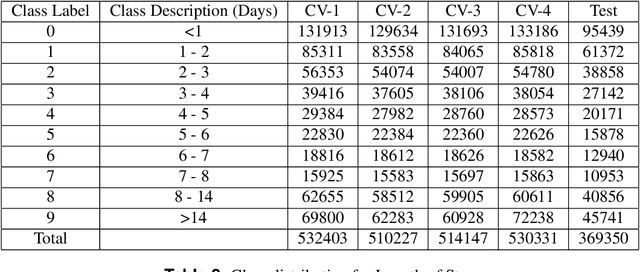
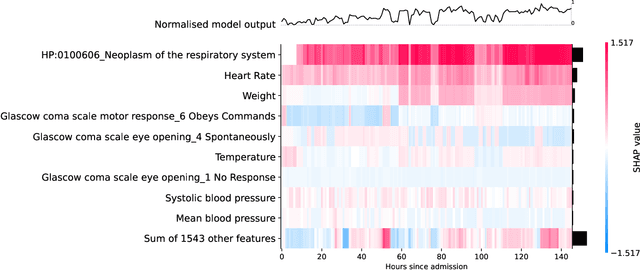
Abstract:Clinical notes contain information not present elsewhere, including drug response and symptoms, all of which are highly important when predicting key outcomes in acute care patients. We propose the automatic annotation of phenotypes from clinical notes as a method to capture essential information to predict outcomes in the Intensive Care Unit (ICU). This information is complementary to typically used vital signs and laboratory test results. We demonstrate and validate our approach conducting experiments on the prediction of in-hospital mortality, physiological decompensation and length of stay in the ICU setting for over 24,000 patients. The prediction models incorporating phenotypic information consistently outperform the baseline models leveraging only vital signs and laboratory test results. Moreover, we conduct a thorough interpretability study, showing that phenotypes provide valuable insights at the patient and cohort levels. Our approach illustrates the viability of using phenotypes to determine outcomes in the ICU.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge