Jingqing Zhang
The Potential and Pitfalls of using a Large Language Model such as ChatGPT or GPT-4 as a Clinical Assistant
Jul 16, 2023
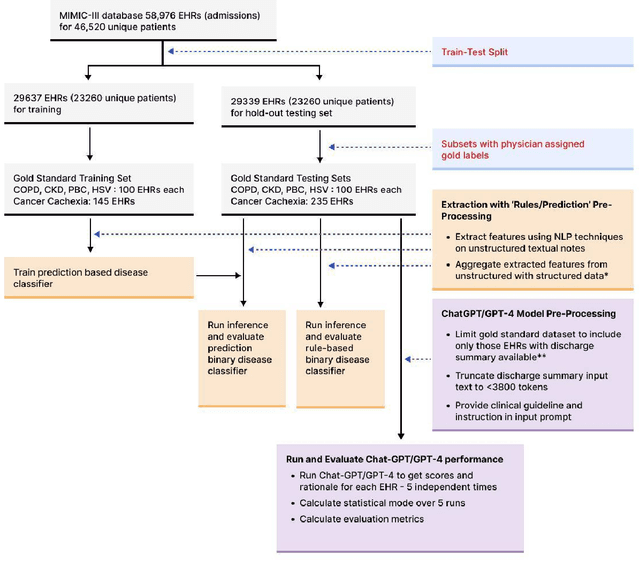
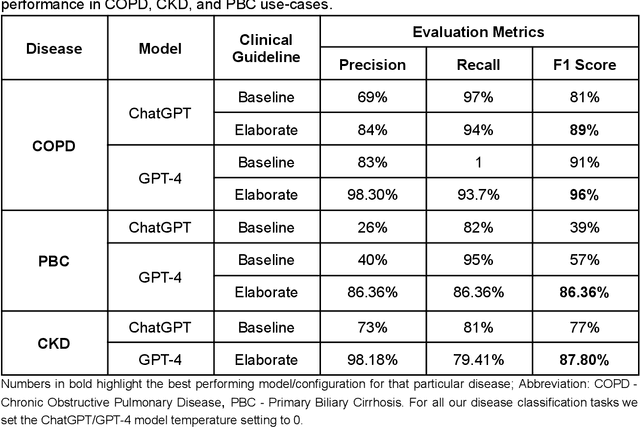
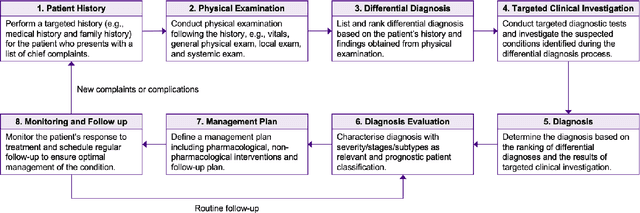
Abstract:Recent studies have demonstrated promising performance of ChatGPT and GPT-4 on several medical domain tasks. However, none have assessed its performance using a large-scale real-world electronic health record database, nor have evaluated its utility in providing clinical diagnostic assistance for patients across a full range of disease presentation. We performed two analyses using ChatGPT and GPT-4, one to identify patients with specific medical diagnoses using a real-world large electronic health record database and the other, in providing diagnostic assistance to healthcare workers in the prospective evaluation of hypothetical patients. Our results show that GPT-4 across disease classification tasks with chain of thought and few-shot prompting can achieve performance as high as 96% F1 scores. For patient assessment, GPT-4 can accurately diagnose three out of four times. However, there were mentions of factually incorrect statements, overlooking crucial medical findings, recommendations for unnecessary investigations and overtreatment. These issues coupled with privacy concerns, make these models currently inadequate for real world clinical use. However, limited data and time needed for prompt engineering in comparison to configuration of conventional machine learning workflows highlight their potential for scalability across healthcare applications.
Medical Scientific Table-to-Text Generation with Human-in-the-Loop under the Data Sparsity Constraint
May 24, 2022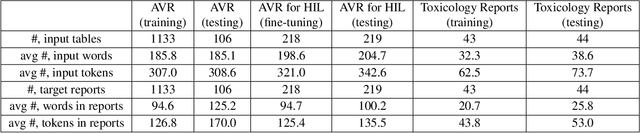
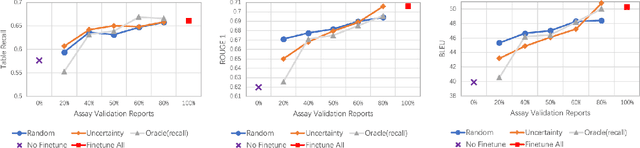
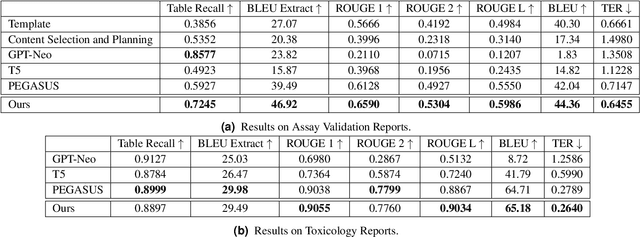
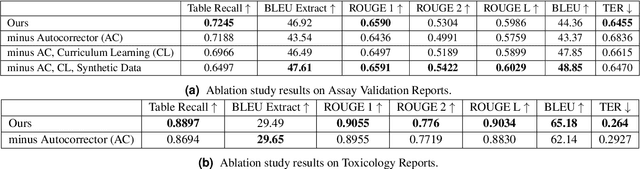
Abstract:Structured (tabular) data in the preclinical and clinical domains contains valuable information about individuals and an efficient table-to-text summarization system can drastically reduce manual efforts to condense this data into reports. However, in practice, the problem is heavily impeded by the data paucity, data sparsity and inability of the state-of-the-art natural language generation models (including T5, PEGASUS and GPT-Neo) to produce accurate and reliable outputs. In this paper, we propose a novel table-to-text approach and tackle these problems with a novel two-step architecture which is enhanced by auto-correction, copy mechanism and synthetic data augmentation. The study shows that the proposed approach selects salient biomedical entities and values from structured data with improved precision (up to 0.13 absolute increase) of copying the tabular values to generate coherent and accurate text for assay validation reports and toxicology reports. Moreover, we also demonstrate a light-weight adaptation of the proposed system to new datasets by fine-tuning with as little as 40\% training examples. The outputs of our model are validated by human experts in the Human-in-the-Loop scenario.
A Scalable Workflow to Build Machine Learning Classifiers with Clinician-in-the-Loop to Identify Patients in Specific Diseases
May 18, 2022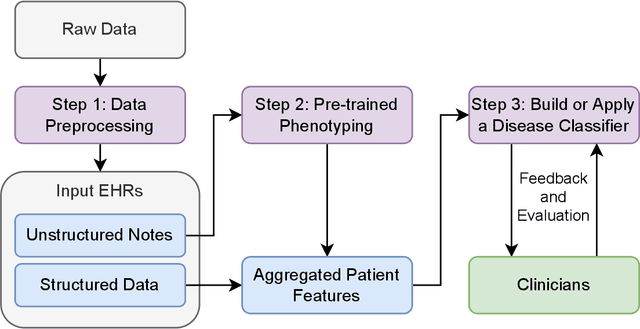

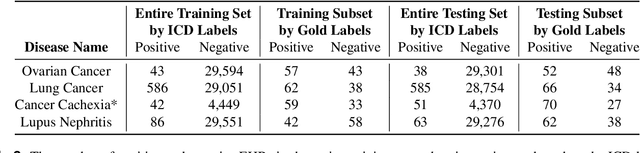
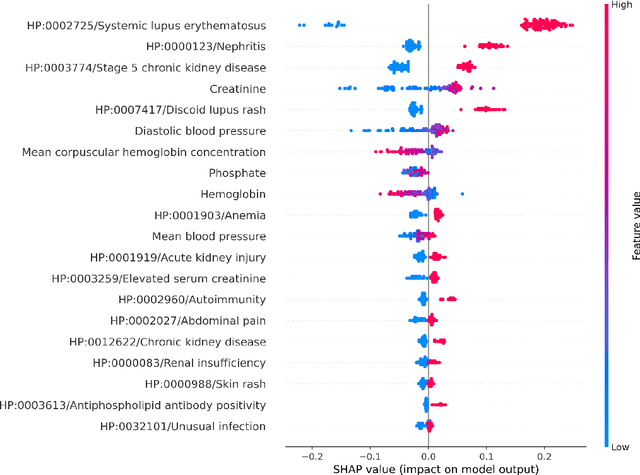
Abstract:Clinicians may rely on medical coding systems such as International Classification of Diseases (ICD) to identify patients with diseases from Electronic Health Records (EHRs). However, due to the lack of detail and specificity as well as a probability of miscoding, recent studies suggest the ICD codes often cannot characterise patients accurately for specific diseases in real clinical practice, and as a result, using them to find patients for studies or trials can result in high failure rates and missing out on uncoded patients. Manual inspection of all patients at scale is not feasible as it is highly costly and slow. This paper proposes a scalable workflow which leverages both structured data and unstructured textual notes from EHRs with techniques including NLP, AutoML and Clinician-in-the-Loop mechanism to build machine learning classifiers to identify patients at scale with given diseases, especially those who might currently be miscoded or missed by ICD codes. Case studies in the MIMIC-III dataset were conducted where the proposed workflow demonstrates a higher classification performance in terms of F1 scores compared to simply using ICD codes on gold testing subset to identify patients with Ovarian Cancer (0.901 vs 0.814), Lung Cancer (0.859 vs 0.828), Cancer Cachexia (0.862 vs 0.650), and Lupus Nephritis (0.959 vs 0.855). Also, the proposed workflow that leverages unstructured notes consistently outperforms the baseline that uses structured data only with an increase of F1 (Ovarian Cancer 0.901 vs 0.719, Lung Cancer 0.859 vs 0.787, Cancer Cachexia 0.862 vs 0.838 and Lupus Nephritis 0.959 vs 0.785). Experiments on the large testing set also demonstrate the proposed workflow can find more patients who are miscoded or missed by ICD codes. Moreover, interpretability studies are also conducted to clinically validate the top impact features of the classifiers.
Unsupervised Numerical Reasoning to Extract Phenotypes from Clinical Text by Leveraging External Knowledge
Apr 19, 2022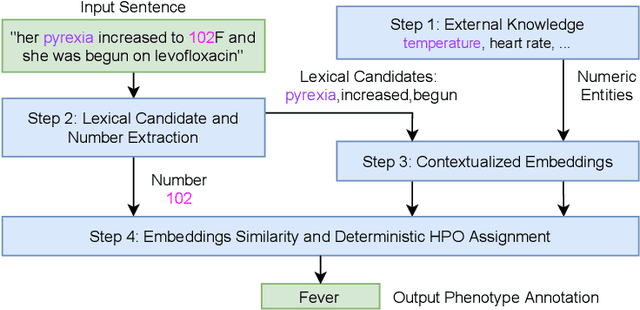
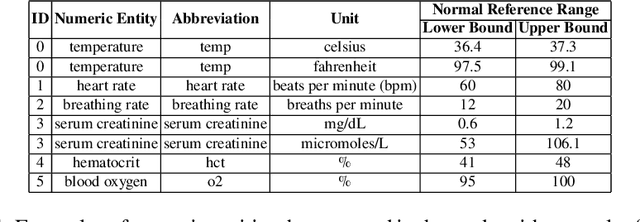
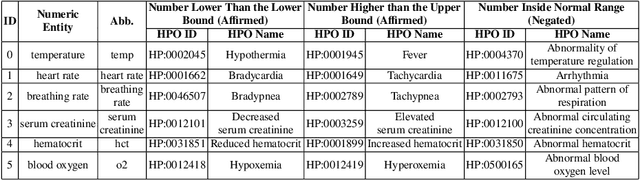
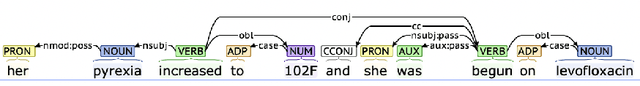
Abstract:Extracting phenotypes from clinical text has been shown to be useful for a variety of clinical use cases such as identifying patients with rare diseases. However, reasoning with numerical values remains challenging for phenotyping in clinical text, for example, temperature 102F representing Fever. Current state-of-the-art phenotyping models are able to detect general phenotypes, but perform poorly when they detect phenotypes requiring numerical reasoning. We present a novel unsupervised methodology leveraging external knowledge and contextualized word embeddings from ClinicalBERT for numerical reasoning in a variety of phenotypic contexts. Comparing against unsupervised benchmarks, it shows a substantial performance improvement with absolute gains on generalized Recall and F1 scores up to 79% and 71%, respectively. In the supervised setting, it also surpasses the performance of alternative approaches with absolute gains on generalized Recall and F1 scores up to 70% and 44%, respectively.
Self-Supervised Detection of Contextual Synonyms in a Multi-Class Setting: Phenotype Annotation Use Case
Sep 04, 2021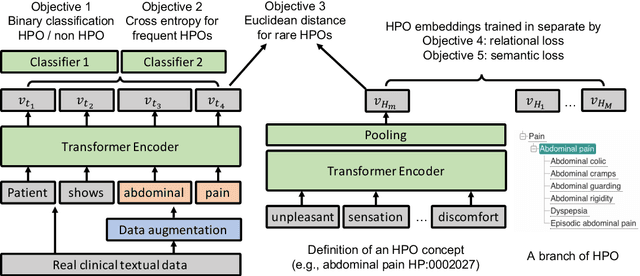
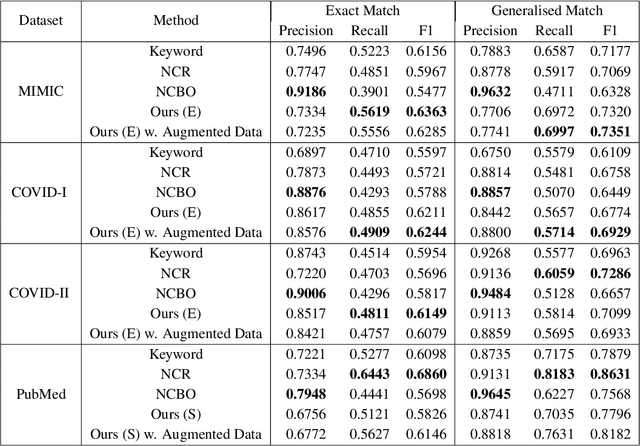
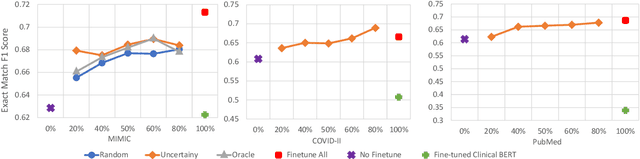
Abstract:Contextualised word embeddings is a powerful tool to detect contextual synonyms. However, most of the current state-of-the-art (SOTA) deep learning concept extraction methods remain supervised and underexploit the potential of the context. In this paper, we propose a self-supervised pre-training approach which is able to detect contextual synonyms of concepts being training on the data created by shallow matching. We apply our methodology in the sparse multi-class setting (over 15,000 concepts) to extract phenotype information from electronic health records. We further investigate data augmentation techniques to address the problem of the class sparsity. Our approach achieves a new SOTA for the unsupervised phenotype concept annotation on clinical text on F1 and Recall outperforming the previous SOTA with a gain of up to 4.5 and 4.0 absolute points, respectively. After fine-tuning with as little as 20\% of the labelled data, we also outperform BioBERT and ClinicalBERT. The extrinsic evaluation on three ICU benchmarks also shows the benefit of using the phenotypes annotated by our model as features.
Clinical Utility of the Automatic Phenotype Annotation in Unstructured Clinical Notes: ICU Use Cases
Jul 24, 2021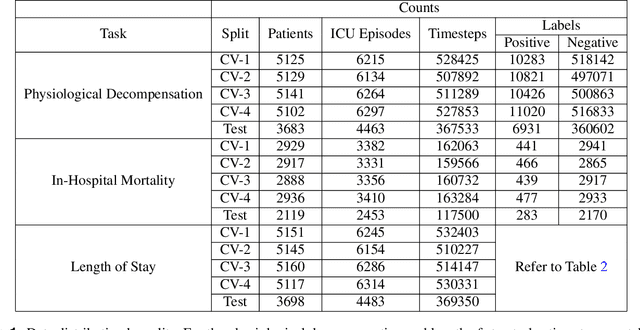
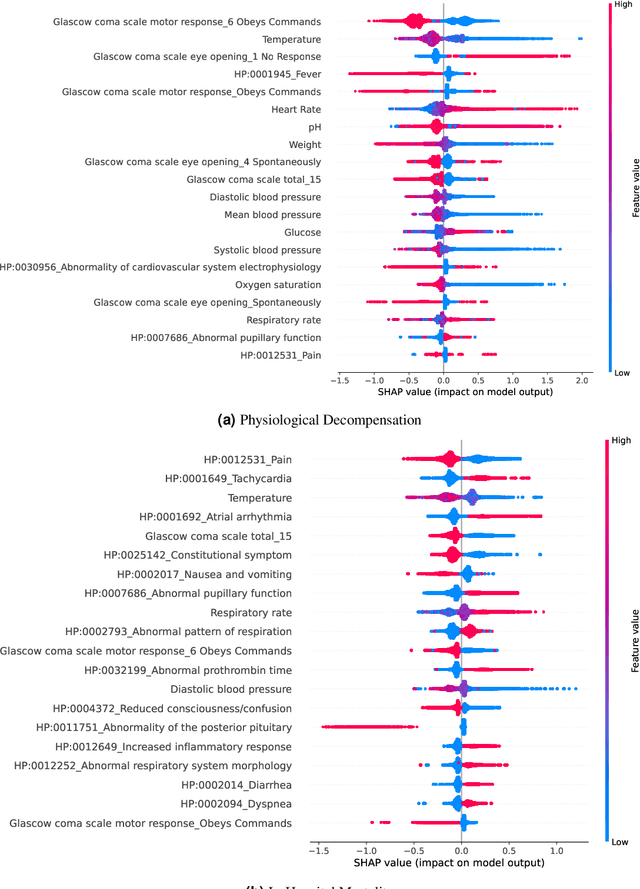
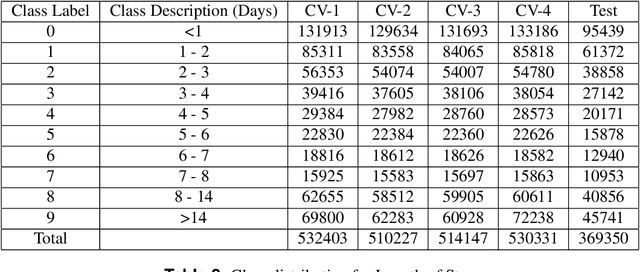
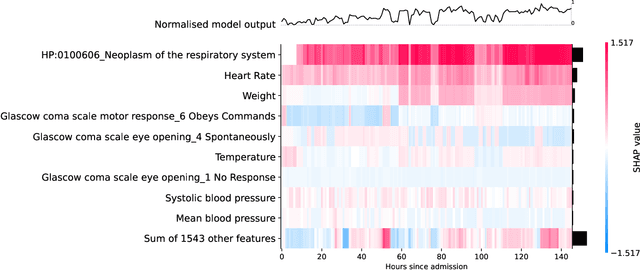
Abstract:Clinical notes contain information not present elsewhere, including drug response and symptoms, all of which are highly important when predicting key outcomes in acute care patients. We propose the automatic annotation of phenotypes from clinical notes as a method to capture essential information to predict outcomes in the Intensive Care Unit (ICU). This information is complementary to typically used vital signs and laboratory test results. We demonstrate and validate our approach conducting experiments on the prediction of in-hospital mortality, physiological decompensation and length of stay in the ICU setting for over 24,000 patients. The prediction models incorporating phenotypic information consistently outperform the baseline models leveraging only vital signs and laboratory test results. Moreover, we conduct a thorough interpretability study, showing that phenotypes provide valuable insights at the patient and cohort levels. Our approach illustrates the viability of using phenotypes to determine outcomes in the ICU.
PEGASUS: Pre-training with Extracted Gap-sentences for Abstractive Summarization
Dec 18, 2019



Abstract:Recent work pre-training Transformers with self-supervised objectives on large text corpora has shown great success when fine-tuned on downstream NLP tasks including text summarization. However, pre-training objectives tailored for abstractive text summarization have not been explored. Furthermore there is a lack of systematic evaluation across diverse domains. In this work, we propose pre-training large Transformer-based encoder-decoder models on massive text corpora with a new self-supervised objective. In PEGASUS, important sentences are removed/masked from an input document and are generated together as one output sequence from the remaining sentences, similar to an extractive summary. We evaluated our best PEGASUS model on 12 downstream summarization tasks spanning news, science, stories, instructions, emails, patents, and legislative bills. Experiments demonstrate it achieves state-of-the-art performance on all 12 downstream datasets measured by ROUGE scores. Our model also shows surprising performance on low-resource summarization, surpassing previous state-of-the-art results on 6 datasets with only 1000 examples.
Unsupervised Annotation of Phenotypic Abnormalities via Semantic Latent Representations on Electronic Health Records
Nov 10, 2019



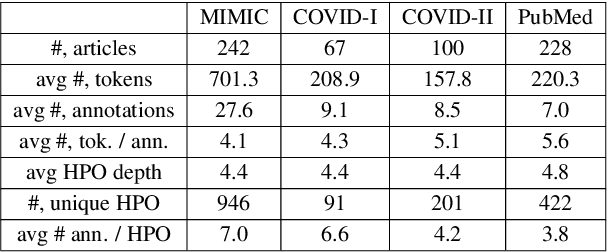
Abstract:The extraction of phenotype information which is naturally contained in electronic health records (EHRs) has been found to be useful in various clinical informatics applications such as disease diagnosis. However, due to imprecise descriptions, lack of gold standards and the demand for efficiency, annotating phenotypic abnormalities on millions of EHR narratives is still challenging. In this work, we propose a novel unsupervised deep learning framework to annotate the phenotypic abnormalities from EHRs via semantic latent representations. The proposed framework takes the advantage of Human Phenotype Ontology (HPO), which is a knowledge base of phenotypic abnormalities, to standardize the annotation results. Experiments have been conducted on 52,722 EHRs from MIMIC-III dataset. Quantitative and qualitative analysis have shown the proposed framework achieves state-of-the-art annotation performance and computational efficiency compared with other methods.
Integrated Multi-omics Analysis Using Variational Autoencoders: Application to Pan-cancer Classification
Aug 17, 2019



Abstract:Different aspects of a clinical sample can be revealed by multiple types of omics data. Integrated analysis of multi-omics data provides a comprehensive view of patients, which has the potential to facilitate more accurate clinical decision making. However, omics data are normally high dimensional with large number of molecular features and relatively small number of available samples with clinical labels. The "dimensionality curse" makes it challenging to train a machine learning model using high dimensional omics data like DNA methylation and gene expression profiles. Here we propose an end-to-end deep learning model called OmiVAE to extract low dimensional features and classify samples from multi-omics data. OmiVAE combines the basic structure of variational autoencoders with a classification network to achieve task-oriented feature extraction and multi-class classification. The training procedure of OmiVAE is comprised of an unsupervised phase without the classifier and a supervised phase with the classifier. During the unsupervised phase, a hierarchical cluster structure of samples can be automatically formed without the need for labels. And in the supervised phase, OmiVAE achieved an average classification accuracy of 97.49% after 10-fold cross-validation among 33 tumour types and normal samples, which shows better performance than other existing methods. The OmiVAE model learned from multi-omics data outperformed that using only one type of omics data, which indicates that the complementary information from different omics datatypes provides useful insights for biomedical tasks like cancer classification.
Integrating Semantic Knowledge to Tackle Zero-shot Text Classification
Mar 29, 2019



Abstract:Insufficient or even unavailable training data of emerging classes is a big challenge of many classification tasks, including text classification. Recognising text documents of classes that have never been seen in the learning stage, so-called zero-shot text classification, is therefore difficult and only limited previous works tackled this problem. In this paper, we propose a two-phase framework together with data augmentation and feature augmentation to solve this problem. Four kinds of semantic knowledge (word embeddings, class descriptions, class hierarchy, and a general knowledge graph) are incorporated into the proposed framework to deal with instances of unseen classes effectively. Experimental results show that each and the combination of the two phases achieve the best overall accuracy compared with baselines and recent approaches in classifying real-world texts under the zero-shot scenario.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge