Tina Eliassi-Rad
When do neural ordinary differential equations generalize on complex networks?
Feb 09, 2026Abstract:Neural ordinary differential equations (neural ODEs) can effectively learn dynamical systems from time series data, but their behavior on graph-structured data remains poorly understood, especially when applied to graphs with different size or structure than encountered during training. We study neural ODEs ($\mathtt{nODE}$s) with vector fields following the Barabási-Barzel form, trained on synthetic data from five common dynamical systems on graphs. Using the $\mathbb{S}^1$-model to generate graphs with realistic and tunable structure, we find that degree heterogeneity and the type of dynamical system are the primary factors in determining $\mathtt{nODE}$s' ability to generalize across graph sizes and properties. This extends to $\mathtt{nODE}$s' ability to capture fixed points and maintain performance amid missing data. Average clustering plays a secondary role in determining $\mathtt{nODE}$ performance. Our findings highlight $\mathtt{nODE}$s as a powerful approach to understanding complex systems but underscore challenges emerging from degree heterogeneity and clustering in realistic graphs.
Hypergraph Representations of scRNA-seq Data for Improved Clustering with Random Walks
Jan 20, 2025Abstract:Analysis of single-cell RNA sequencing data is often conducted through network projections such as coexpression networks, primarily due to the abundant availability of network analysis tools for downstream tasks. However, this approach has several limitations: loss of higher-order information, inefficient data representation caused by converting a sparse dataset to a fully connected network, and overestimation of coexpression due to zero-inflation. To address these limitations, we propose conceptualizing scRNA-seq expression data as hypergraphs, which are generalized graphs in which the hyperedges can connect more than two vertices. In the context of scRNA-seq data, the hypergraph nodes represent cells and the edges represent genes. Each hyperedge connects all cells where its corresponding gene is actively expressed and records the expression of the gene across different cells. This hypergraph conceptualization enables us to explore multi-way relationships beyond the pairwise interactions in coexpression networks without loss of information. We propose two novel clustering methods: (1) the Dual-Importance Preference Hypergraph Walk (DIPHW) and (2) the Coexpression and Memory-Integrated Dual-Importance Preference Hypergraph Walk (CoMem-DIPHW). They outperform established methods on both simulated and real scRNA-seq datasets. The improvement brought by our proposed methods is especially significant when data modularity is weak. Furthermore, CoMem-DIPHW incorporates the gene coexpression network, cell coexpression network, and the cell-gene expression hypergraph from the single-cell abundance counts data altogether for embedding computation. This approach accounts for both the local level information from single-cell level gene expression and the global level information from the pairwise similarity in the two coexpression networks.
A Misclassification Network-Based Method for Comparative Genomic Analysis
Dec 09, 2024



Abstract:Classifying genome sequences based on metadata has been an active area of research in comparative genomics for decades with many important applications across the life sciences. Established methods for classifying genomes can be broadly grouped into sequence alignment-based and alignment-free models. Conventional alignment-based models rely on genome similarity measures calculated based on local sequence alignments or consistent ordering among sequences. However, such methods are computationally expensive when dealing with large ensembles of even moderately sized genomes. In contrast, alignment-free (AF) approaches measure genome similarity based on summary statistics in an unsupervised setting and are efficient enough to analyze large datasets. However, both alignment-based and AF methods typically assume fixed scoring rubrics that lack the flexibility to assign varying importance to different parts of the sequences based on prior knowledge. In this study, we integrate AI and network science approaches to develop a comparative genomic analysis framework that addresses these limitations. Our approach, termed the Genome Misclassification Network Analysis (GMNA), simultaneously leverages misclassified instances, a learned scoring rubric, and label information to classify genomes based on associated metadata and better understand potential drivers of misclassification. We evaluate the utility of the GMNA using Naive Bayes and convolutional neural network models, supplemented by additional experiments with transformer-based models, to construct SARS-CoV-2 sampling location classifiers using over 500,000 viral genome sequences and study the resulting network of misclassifications. We demonstrate the global health potential of the GMNA by leveraging the SARS-CoV-2 genome misclassification networks to investigate the role human mobility played in structuring geographic clustering of SARS-CoV-2.
REGE: A Method for Incorporating Uncertainty in Graph Embeddings
Dec 07, 2024



Abstract:Machine learning models for graphs in real-world applications are prone to two primary types of uncertainty: (1) those that arise from incomplete and noisy data and (2) those that arise from uncertainty of the model in its output. These sources of uncertainty are not mutually exclusive. Additionally, models are susceptible to targeted adversarial attacks, which exacerbate both of these uncertainties. In this work, we introduce Radius Enhanced Graph Embeddings (REGE), an approach that measures and incorporates uncertainty in data to produce graph embeddings with radius values that represent the uncertainty of the model's output. REGE employs curriculum learning to incorporate data uncertainty and conformal learning to address the uncertainty in the model's output. In our experiments, we show that REGE's graph embeddings perform better under adversarial attacks by an average of 1.5% (accuracy) against state-of-the-art methods.
Generating Human Understandable Explanations for Node Embeddings
Jun 11, 2024



Abstract:Node embedding algorithms produce low-dimensional latent representations of nodes in a graph. These embeddings are often used for downstream tasks, such as node classification and link prediction. In this paper, we investigate the following two questions: (Q1) Can we explain each embedding dimension with human-understandable graph features (e.g. degree, clustering coefficient and PageRank). (Q2) How can we modify existing node embedding algorithms to produce embeddings that can be easily explained by human-understandable graph features? We find that the answer to Q1 is yes and introduce a new framework called XM (short for eXplain eMbedding) to answer Q2. A key aspect of XM involves minimizing the nuclear norm of the generated explanations. We show that by minimizing the nuclear norm, we minimize the lower bound on the entropy of the generated explanations. We test XM on a variety of real-world graphs and show that XM not only preserves the performance of existing node embedding methods, but also enhances their explainability.
Re-visiting Skip-Gram Negative Sampling: Dimension Regularization for More Efficient Dissimilarity Preservation in Graph Embeddings
Apr 30, 2024



Abstract:A wide range of graph embedding objectives decompose into two components: one that attracts the embeddings of nodes that are perceived as similar, and another that repels embeddings of nodes that are perceived as dissimilar. Because real-world graphs are sparse and the number of dissimilar pairs grows quadratically with the number of nodes, Skip-Gram Negative Sampling (SGNS) has emerged as a popular and efficient repulsion approach. SGNS repels each node from a sample of dissimilar nodes, as opposed to all dissimilar nodes. In this work, we show that node-wise repulsion is, in aggregate, an approximate re-centering of the node embedding dimensions. Such dimension operations are much more scalable than node operations. The dimension approach, in addition to being more efficient, yields a simpler geometric interpretation of the repulsion. Our result extends findings from the self-supervised learning literature to the skip-gram model, establishing a connection between skip-gram node contrast and dimension regularization. We show that in the limit of large graphs, under mild regularity conditions, the original node repulsion objective converges to optimization with dimension regularization. We use this observation to propose an algorithm augmentation framework that speeds up any existing algorithm, supervised or unsupervised, using SGNS. The framework prioritizes node attraction and replaces SGNS with dimension regularization. We instantiate this generic framework for LINE and node2vec and show that the augmented algorithms preserve downstream performance while dramatically increasing efficiency.
GRASP: Accelerating Shortest Path Attacks via Graph Attention
Oct 23, 2023Abstract:Recent advances in machine learning (ML) have shown promise in aiding and accelerating classical combinatorial optimization algorithms. ML-based speed ups that aim to learn in an end to end manner (i.e., directly output the solution) tend to trade off run time with solution quality. Therefore, solutions that are able to accelerate existing solvers while maintaining their performance guarantees, are of great interest. We consider an APX-hard problem, where an adversary aims to attack shortest paths in a graph by removing the minimum number of edges. We propose the GRASP algorithm: Graph Attention Accelerated Shortest Path Attack, an ML aided optimization algorithm that achieves run times up to 10x faster, while maintaining the quality of solution generated. GRASP uses a graph attention network to identify a smaller subgraph containing the combinatorial solution, thus effectively reducing the input problem size. Additionally, we demonstrate how careful representation of the input graph, including node features that correlate well with the optimization task, can highlight important structure in the optimization solution.
When Collaborative Filtering is not Collaborative: Unfairness of PCA for Recommendations
Oct 15, 2023



Abstract:We study the fairness of dimensionality reduction methods for recommendations. We focus on the established method of principal component analysis (PCA), which identifies latent components and produces a low-rank approximation via the leading components while discarding the trailing components. Prior works have defined notions of "fair PCA"; however, these definitions do not answer the following question: what makes PCA unfair? We identify two underlying mechanisms of PCA that induce unfairness at the item level. The first negatively impacts less popular items, due to the fact that less popular items rely on trailing latent components to recover their values. The second negatively impacts the highly popular items, since the leading PCA components specialize in individual popular items instead of capturing similarities between items. To address these issues, we develop a polynomial-time algorithm, Item-Weighted PCA, a modification of PCA that uses item-specific weights in the objective. On a stylized class of matrices, we prove that Item-Weighted PCA using a specific set of weights minimizes a popularity-normalized error metric. Our evaluations on real-world datasets show that Item-Weighted PCA not only improves overall recommendation quality by up to $0.1$ item-level AUC-ROC but also improves on both popular and less popular items.
Graph-SCP: Accelerating Set Cover Problems with Graph Neural Networks
Oct 12, 2023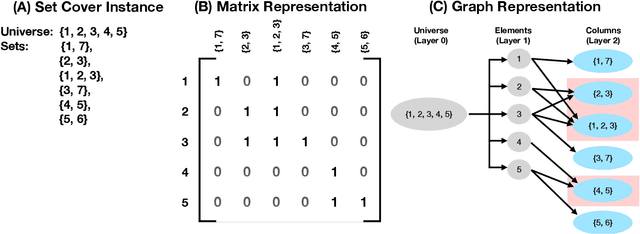
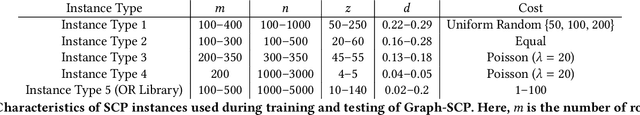
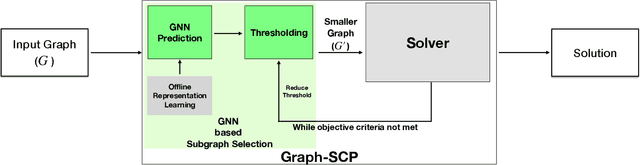
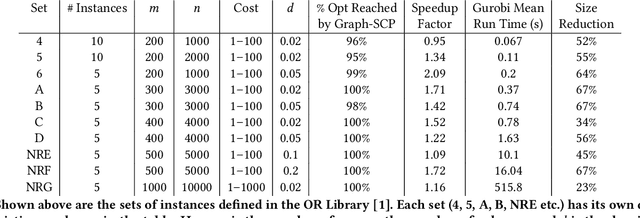
Abstract:Machine learning (ML) approaches are increasingly being used to accelerate combinatorial optimization (CO) problems. We look specifically at the Set Cover Problem (SCP) and propose Graph-SCP, a graph neural network method that can augment existing optimization solvers by learning to identify a much smaller sub-problem that contains the solution space. We evaluate the performance of Graph-SCP on synthetic weighted and unweighted SCP instances with diverse problem characteristics and complexities, and on instances from the OR Library, a canonical benchmark for SCP. We show that Graph-SCP reduces the problem size by 30-70% and achieves run time speedups up to~25x when compared to commercial solvers (Gurobi). Given a desired optimality threshold, Graph-SCP will improve upon it or even achieve 100% optimality. This is in contrast to fast greedy solutions that significantly compromise solution quality to achieve guaranteed polynomial run time. Graph-SCP can generalize to larger problem sizes and can be used with other conventional or ML-augmented CO solvers to lead to potential additional run time improvement.
Disentangling Node Attributes from Graph Topology for Improved Generalizability in Link Prediction
Jul 17, 2023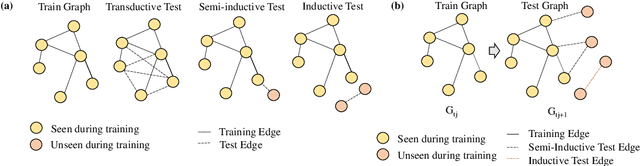
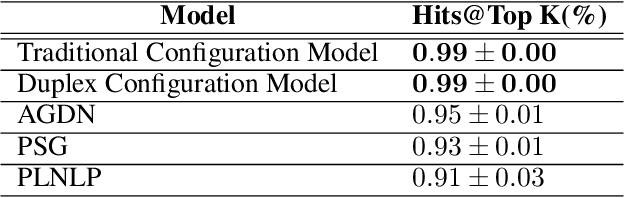
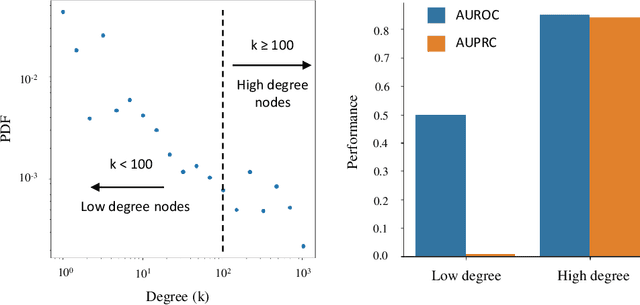
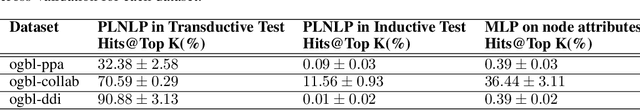
Abstract:Link prediction is a crucial task in graph machine learning with diverse applications. We explore the interplay between node attributes and graph topology and demonstrate that incorporating pre-trained node attributes improves the generalization power of link prediction models. Our proposed method, UPNA (Unsupervised Pre-training of Node Attributes), solves the inductive link prediction problem by learning a function that takes a pair of node attributes and predicts the probability of an edge, as opposed to Graph Neural Networks (GNN), which can be prone to topological shortcuts in graphs with power-law degree distribution. In this manner, UPNA learns a significant part of the latent graph generation mechanism since the learned function can be used to add incoming nodes to a growing graph. By leveraging pre-trained node attributes, we overcome observational bias and make meaningful predictions about unobserved nodes, surpassing state-of-the-art performance (3X to 34X improvement on benchmark datasets). UPNA can be applied to various pairwise learning tasks and integrated with existing link prediction models to enhance their generalizability and bolster graph generative models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge