Giulia Menichetti
Informatics for Food Processing
May 20, 2025Abstract:This chapter explores the evolution, classification, and health implications of food processing, while emphasizing the transformative role of machine learning, artificial intelligence (AI), and data science in advancing food informatics. It begins with a historical overview and a critical review of traditional classification frameworks such as NOVA, Nutri-Score, and SIGA, highlighting their strengths and limitations, particularly the subjectivity and reproducibility challenges that hinder epidemiological research and public policy. To address these issues, the chapter presents novel computational approaches, including FoodProX, a random forest model trained on nutrient composition data to infer processing levels and generate a continuous FPro score. It also explores how large language models like BERT and BioBERT can semantically embed food descriptions and ingredient lists for predictive tasks, even in the presence of missing data. A key contribution of the chapter is a novel case study using the Open Food Facts database, showcasing how multimodal AI models can integrate structured and unstructured data to classify foods at scale, offering a new paradigm for food processing assessment in public health and research.
Disentangling Node Attributes from Graph Topology for Improved Generalizability in Link Prediction
Jul 17, 2023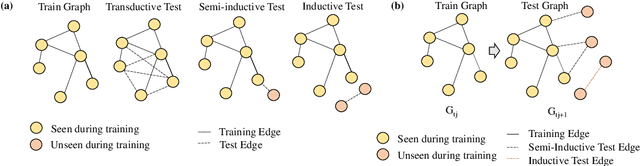
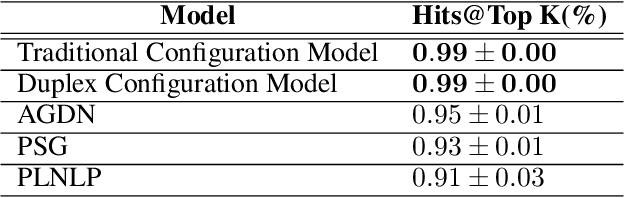
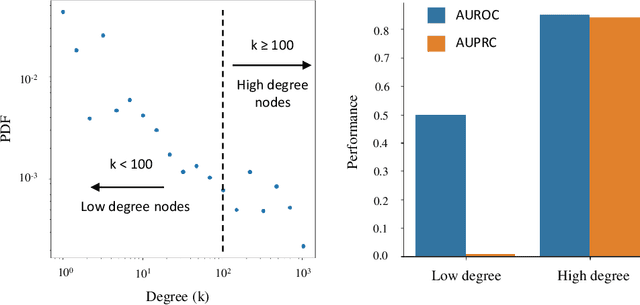
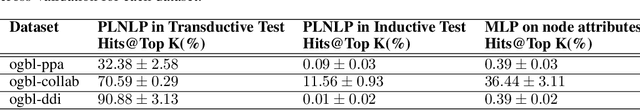
Abstract:Link prediction is a crucial task in graph machine learning with diverse applications. We explore the interplay between node attributes and graph topology and demonstrate that incorporating pre-trained node attributes improves the generalization power of link prediction models. Our proposed method, UPNA (Unsupervised Pre-training of Node Attributes), solves the inductive link prediction problem by learning a function that takes a pair of node attributes and predicts the probability of an edge, as opposed to Graph Neural Networks (GNN), which can be prone to topological shortcuts in graphs with power-law degree distribution. In this manner, UPNA learns a significant part of the latent graph generation mechanism since the learned function can be used to add incoming nodes to a growing graph. By leveraging pre-trained node attributes, we overcome observational bias and make meaningful predictions about unobserved nodes, surpassing state-of-the-art performance (3X to 34X improvement on benchmark datasets). UPNA can be applied to various pairwise learning tasks and integrated with existing link prediction models to enhance their generalizability and bolster graph generative models.
AI-Bind: Improving Binding Predictions for Novel Protein Targets and Ligands
Dec 28, 2021
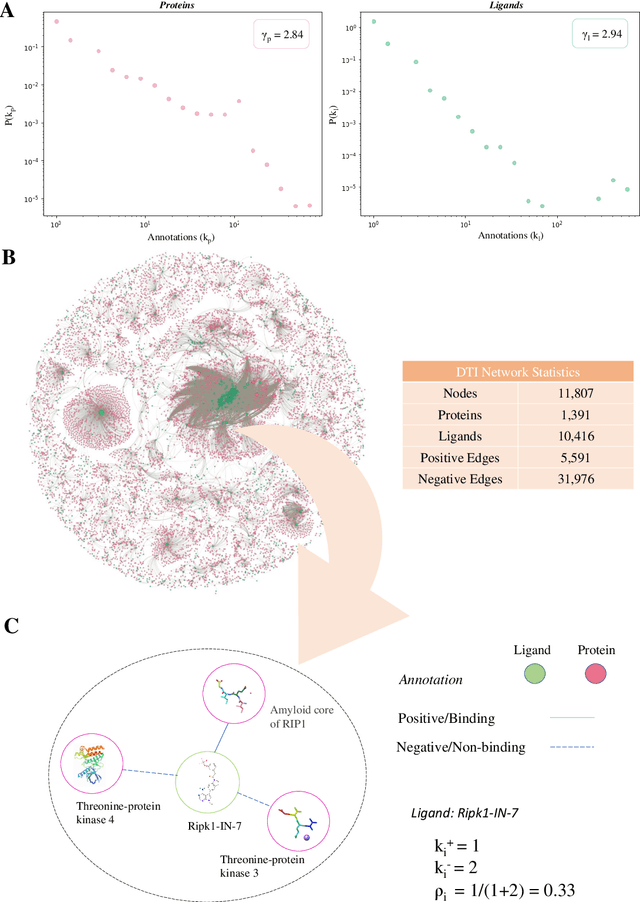

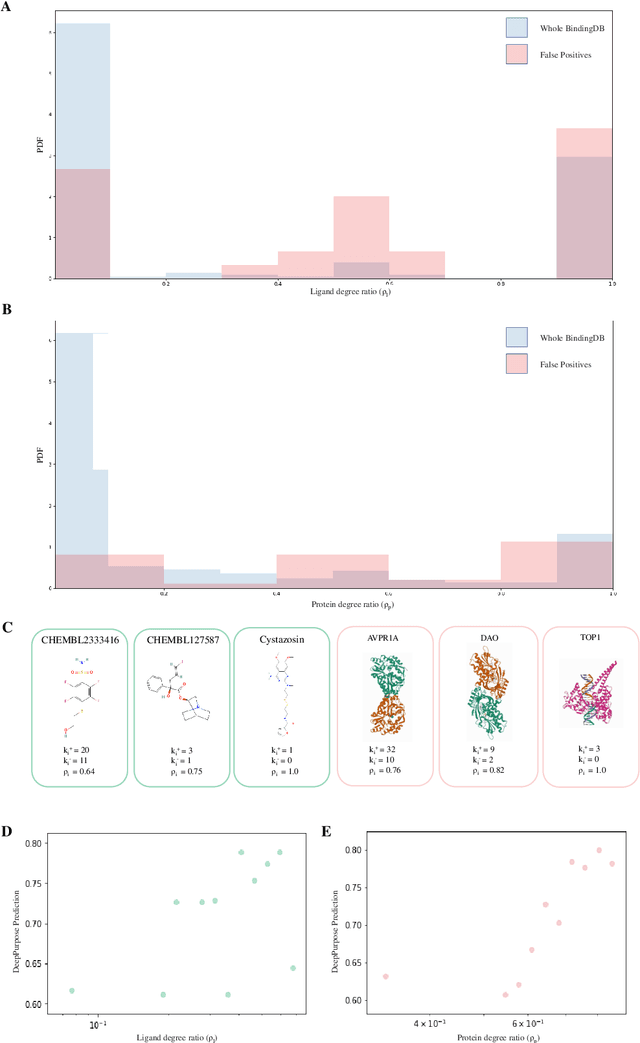
Abstract:Identifying novel drug-target interactions (DTI) is a critical and rate limiting step in drug discovery. While deep learning models have been proposed to accelerate the identification process, we show that state-of-the-art models fail to generalize to novel (i.e., never-before-seen) structures. We first unveil the mechanisms responsible for this shortcoming, demonstrating how models rely on shortcuts that leverage the topology of the protein-ligand bipartite network, rather than learning the node features. Then, we introduce AI-Bind, a pipeline that combines network-based sampling strategies with unsupervised pre-training, allowing us to limit the annotation imbalance and improve binding predictions for novel proteins and ligands. We illustrate the value of AI-Bind by predicting drugs and natural compounds with binding affinity to SARS-CoV-2 viral proteins and the associated human proteins. We also validate these predictions via auto-docking simulations and comparison with recent experimental evidence. Overall, AI-Bind offers a powerful high-throughput approach to identify drug-target combinations, with the potential of becoming a powerful tool in drug discovery.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge