Zohair Shafi
DeepWeightFlow: Re-Basined Flow Matching for Generating Neural Network Weights
Jan 08, 2026Abstract:Building efficient and effective generative models for neural network weights has been a research focus of significant interest that faces challenges posed by the high-dimensional weight spaces of modern neural networks and their symmetries. Several prior generative models are limited to generating partial neural network weights, particularly for larger models, such as ResNet and ViT. Those that do generate complete weights struggle with generation speed or require finetuning of the generated models. In this work, we present DeepWeightFlow, a Flow Matching model that operates directly in weight space to generate diverse and high-accuracy neural network weights for a variety of architectures, neural network sizes, and data modalities. The neural networks generated by DeepWeightFlow do not require fine-tuning to perform well and can scale to large networks. We apply Git Re-Basin and TransFusion for neural network canonicalization in the context of generative weight models to account for the impact of neural network permutation symmetries and to improve generation efficiency for larger model sizes. The generated networks excel at transfer learning, and ensembles of hundreds of neural networks can be generated in minutes, far exceeding the efficiency of diffusion-based methods. DeepWeightFlow models pave the way for more efficient and scalable generation of diverse sets of neural networks.
REGE: A Method for Incorporating Uncertainty in Graph Embeddings
Dec 07, 2024



Abstract:Machine learning models for graphs in real-world applications are prone to two primary types of uncertainty: (1) those that arise from incomplete and noisy data and (2) those that arise from uncertainty of the model in its output. These sources of uncertainty are not mutually exclusive. Additionally, models are susceptible to targeted adversarial attacks, which exacerbate both of these uncertainties. In this work, we introduce Radius Enhanced Graph Embeddings (REGE), an approach that measures and incorporates uncertainty in data to produce graph embeddings with radius values that represent the uncertainty of the model's output. REGE employs curriculum learning to incorporate data uncertainty and conformal learning to address the uncertainty in the model's output. In our experiments, we show that REGE's graph embeddings perform better under adversarial attacks by an average of 1.5% (accuracy) against state-of-the-art methods.
Generating Human Understandable Explanations for Node Embeddings
Jun 11, 2024



Abstract:Node embedding algorithms produce low-dimensional latent representations of nodes in a graph. These embeddings are often used for downstream tasks, such as node classification and link prediction. In this paper, we investigate the following two questions: (Q1) Can we explain each embedding dimension with human-understandable graph features (e.g. degree, clustering coefficient and PageRank). (Q2) How can we modify existing node embedding algorithms to produce embeddings that can be easily explained by human-understandable graph features? We find that the answer to Q1 is yes and introduce a new framework called XM (short for eXplain eMbedding) to answer Q2. A key aspect of XM involves minimizing the nuclear norm of the generated explanations. We show that by minimizing the nuclear norm, we minimize the lower bound on the entropy of the generated explanations. We test XM on a variety of real-world graphs and show that XM not only preserves the performance of existing node embedding methods, but also enhances their explainability.
GRASP: Accelerating Shortest Path Attacks via Graph Attention
Oct 23, 2023Abstract:Recent advances in machine learning (ML) have shown promise in aiding and accelerating classical combinatorial optimization algorithms. ML-based speed ups that aim to learn in an end to end manner (i.e., directly output the solution) tend to trade off run time with solution quality. Therefore, solutions that are able to accelerate existing solvers while maintaining their performance guarantees, are of great interest. We consider an APX-hard problem, where an adversary aims to attack shortest paths in a graph by removing the minimum number of edges. We propose the GRASP algorithm: Graph Attention Accelerated Shortest Path Attack, an ML aided optimization algorithm that achieves run times up to 10x faster, while maintaining the quality of solution generated. GRASP uses a graph attention network to identify a smaller subgraph containing the combinatorial solution, thus effectively reducing the input problem size. Additionally, we demonstrate how careful representation of the input graph, including node features that correlate well with the optimization task, can highlight important structure in the optimization solution.
Graph-SCP: Accelerating Set Cover Problems with Graph Neural Networks
Oct 12, 2023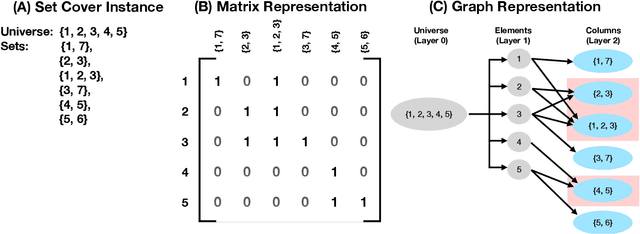
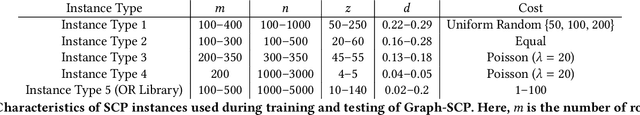
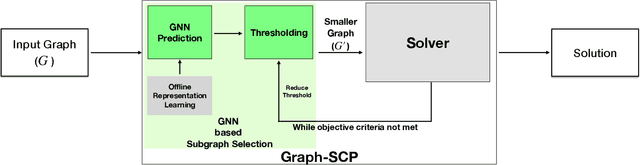
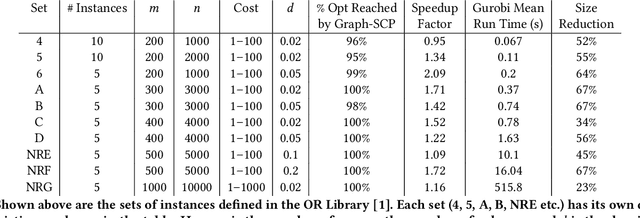
Abstract:Machine learning (ML) approaches are increasingly being used to accelerate combinatorial optimization (CO) problems. We look specifically at the Set Cover Problem (SCP) and propose Graph-SCP, a graph neural network method that can augment existing optimization solvers by learning to identify a much smaller sub-problem that contains the solution space. We evaluate the performance of Graph-SCP on synthetic weighted and unweighted SCP instances with diverse problem characteristics and complexities, and on instances from the OR Library, a canonical benchmark for SCP. We show that Graph-SCP reduces the problem size by 30-70% and achieves run time speedups up to~25x when compared to commercial solvers (Gurobi). Given a desired optimality threshold, Graph-SCP will improve upon it or even achieve 100% optimality. This is in contrast to fast greedy solutions that significantly compromise solution quality to achieve guaranteed polynomial run time. Graph-SCP can generalize to larger problem sizes and can be used with other conventional or ML-augmented CO solvers to lead to potential additional run time improvement.
AI-Bind: Improving Binding Predictions for Novel Protein Targets and Ligands
Dec 28, 2021
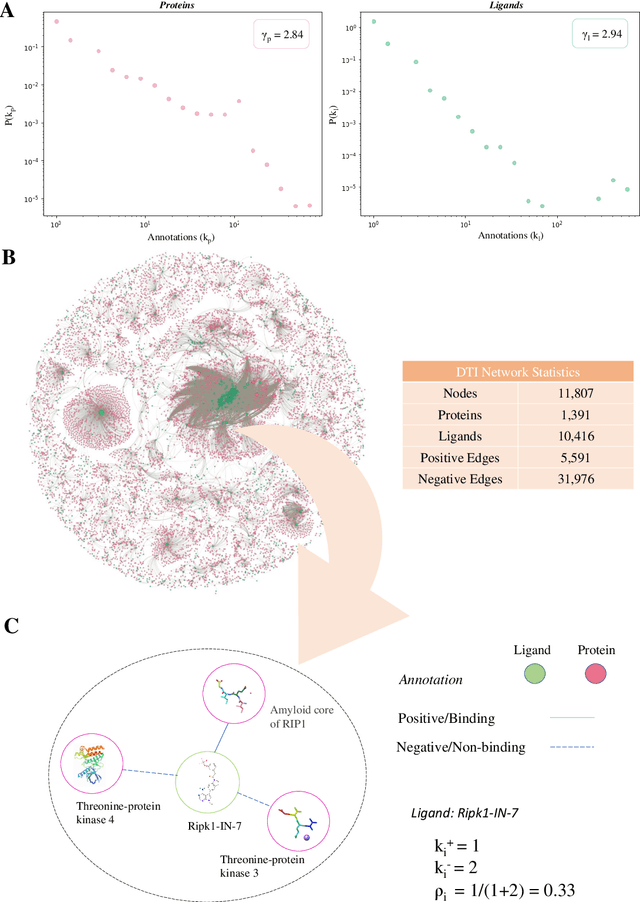

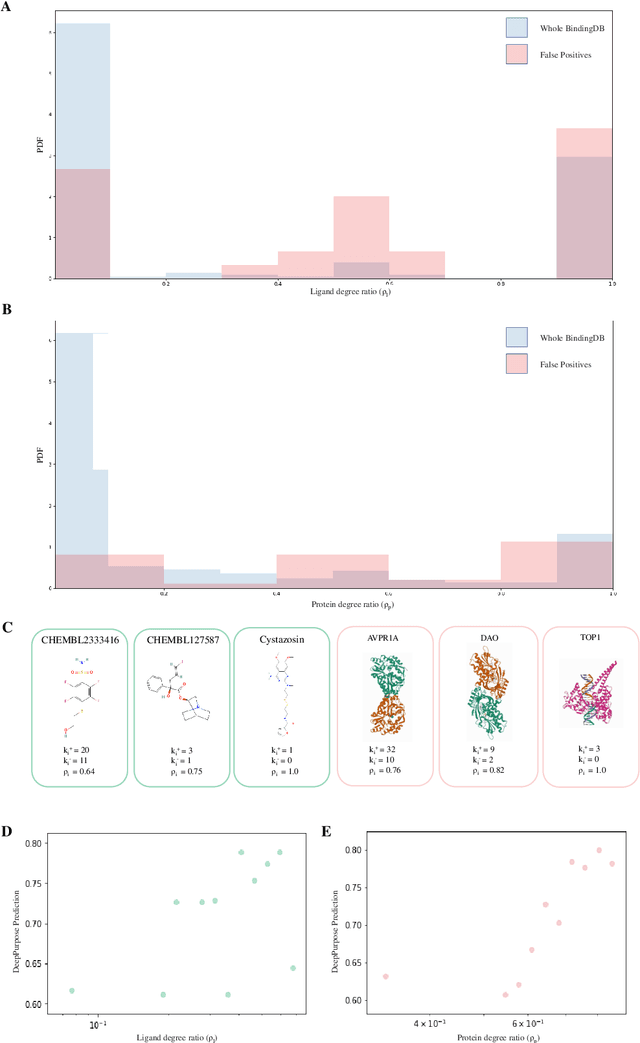
Abstract:Identifying novel drug-target interactions (DTI) is a critical and rate limiting step in drug discovery. While deep learning models have been proposed to accelerate the identification process, we show that state-of-the-art models fail to generalize to novel (i.e., never-before-seen) structures. We first unveil the mechanisms responsible for this shortcoming, demonstrating how models rely on shortcuts that leverage the topology of the protein-ligand bipartite network, rather than learning the node features. Then, we introduce AI-Bind, a pipeline that combines network-based sampling strategies with unsupervised pre-training, allowing us to limit the annotation imbalance and improve binding predictions for novel proteins and ligands. We illustrate the value of AI-Bind by predicting drugs and natural compounds with binding affinity to SARS-CoV-2 viral proteins and the associated human proteins. We also validate these predictions via auto-docking simulations and comparison with recent experimental evidence. Overall, AI-Bind offers a powerful high-throughput approach to identify drug-target combinations, with the potential of becoming a powerful tool in drug discovery.
RAWLSNET: Altering Bayesian Networks to Encode Rawlsian Fair Equality of Opportunity
Mar 16, 2021



Abstract:We present RAWLSNET, a system for altering Bayesian Network (BN) models to satisfy the Rawlsian principle of fair equality of opportunity (FEO). RAWLSNET's BN models generate aspirational data distributions: data generated to reflect an ideally fair, FEO-satisfying society. FEO states that everyone with the same talent and willingness to use it should have the same chance of achieving advantageous social positions (e.g., employment), regardless of their background circumstances (e.g., socioeconomic status). Satisfying FEO requires alterations to social structures such as school assignments. Our paper describes RAWLSNET, a method which takes as input a BN representation of an FEO application and alters the BN's parameters so as to satisfy FEO when possible, and minimize deviation from FEO otherwise. We also offer guidance for applying RAWLSNET, including on recognizing proper applications of FEO. We demonstrate the use of our system with publicly available data sets. RAWLSNET's altered BNs offer the novel capability of generating aspirational data for FEO-relevant tasks. Aspirational data are free from the biases of real-world data, and thus are useful for recognizing and detecting sources of unfairness in machine learning algorithms besides biased data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge