Tawsifur Rahman
Addressing cognitive bias in medical language models
Feb 20, 2024



Abstract:There is increasing interest in the application large language models (LLMs) to the medical field, in part because of their impressive performance on medical exam questions. While promising, exam questions do not reflect the complexity of real patient-doctor interactions. In reality, physicians' decisions are shaped by many complex factors, such as patient compliance, personal experience, ethical beliefs, and cognitive bias. Taking a step toward understanding this, our hypothesis posits that when LLMs are confronted with clinical questions containing cognitive biases, they will yield significantly less accurate responses compared to the same questions presented without such biases. In this study, we developed BiasMedQA, a benchmark for evaluating cognitive biases in LLMs applied to medical tasks. Using BiasMedQA we evaluated six LLMs, namely GPT-4, Mixtral-8x70B, GPT-3.5, PaLM-2, Llama 2 70B-chat, and the medically specialized PMC Llama 13B. We tested these models on 1,273 questions from the US Medical Licensing Exam (USMLE) Steps 1, 2, and 3, modified to replicate common clinically-relevant cognitive biases. Our analysis revealed varying effects for biases on these LLMs, with GPT-4 standing out for its resilience to bias, in contrast to Llama 2 70B-chat and PMC Llama 13B, which were disproportionately affected by cognitive bias. Our findings highlight the critical need for bias mitigation in the development of medical LLMs, pointing towards safer and more reliable applications in healthcare.
BIO-CXRNET: A Robust Multimodal Stacking Machine Learning Technique for Mortality Risk Prediction of COVID-19 Patients using Chest X-Ray Images and Clinical Data
Jun 15, 2022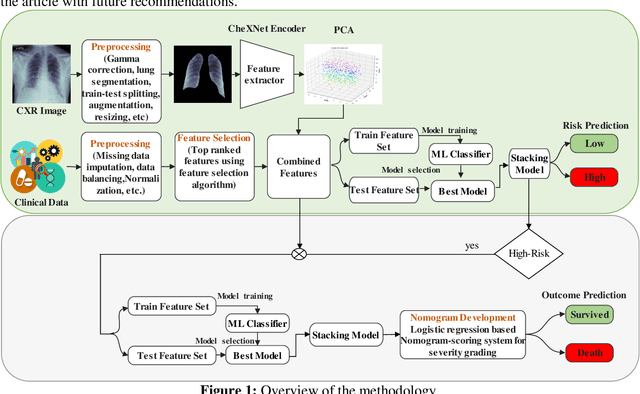
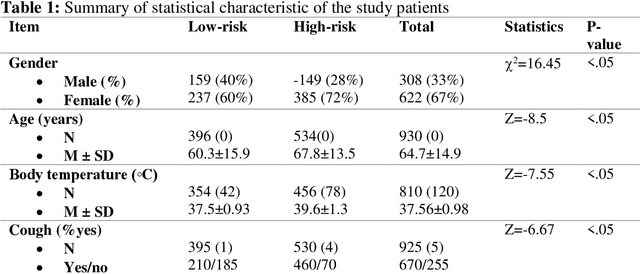
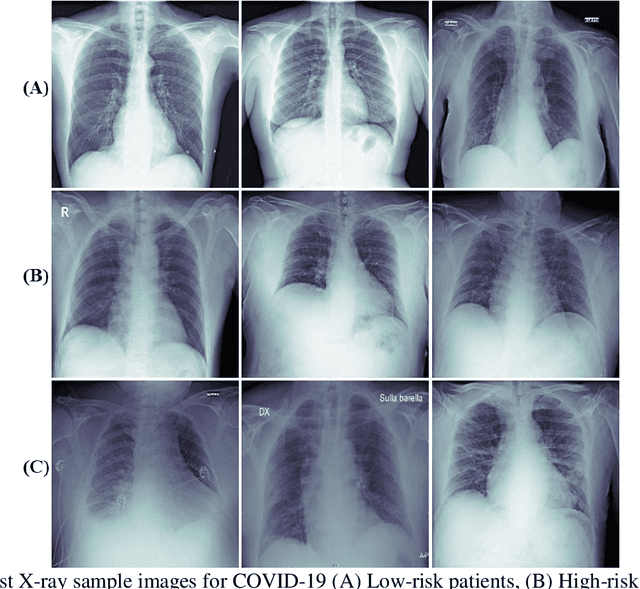
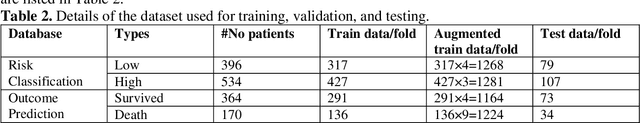
Abstract:Fast and accurate detection of the disease can significantly help in reducing the strain on the healthcare facility of any country to reduce the mortality during any pandemic. The goal of this work is to create a multimodal system using a novel machine learning framework that uses both Chest X-ray (CXR) images and clinical data to predict severity in COVID-19 patients. In addition, the study presents a nomogram-based scoring technique for predicting the likelihood of death in high-risk patients. This study uses 25 biomarkers and CXR images in predicting the risk in 930 COVID-19 patients admitted during the first wave of COVID-19 (March-June 2020) in Italy. The proposed multimodal stacking technique produced the precision, sensitivity, and F1-score, of 89.03%, 90.44%, and 89.03%, respectively to identify low or high-risk patients. This multimodal approach improved the accuracy by 6% in comparison to the CXR image or clinical data alone. Finally, nomogram scoring system using multivariate logistic regression -- was used to stratify the mortality risk among the high-risk patients identified in the first stage. Lactate Dehydrogenase (LDH), O2 percentage, White Blood Cells (WBC) Count, Age, and C-reactive protein (CRP) were identified as useful predictor using random forest feature selection model. Five predictors parameters and a CXR image based nomogram score was developed for quantifying the probability of death and categorizing them into two risk groups: survived (<50%), and death (>=50%), respectively. The multi-modal technique was able to predict the death probability of high-risk patients with an F1 score of 92.88 %. The area under the curves for the development and validation cohorts are 0.981 and 0.939, respectively.
Blind ECG Restoration by Operational Cycle-GANs
Jan 29, 2022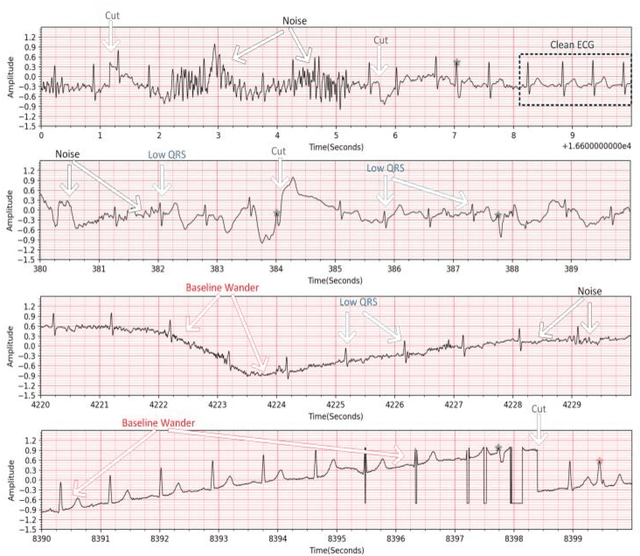
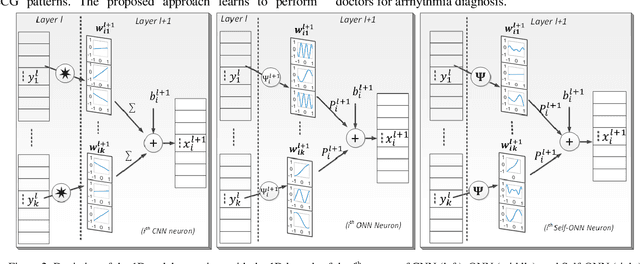
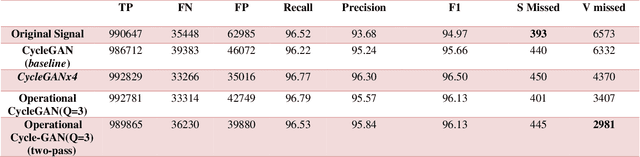
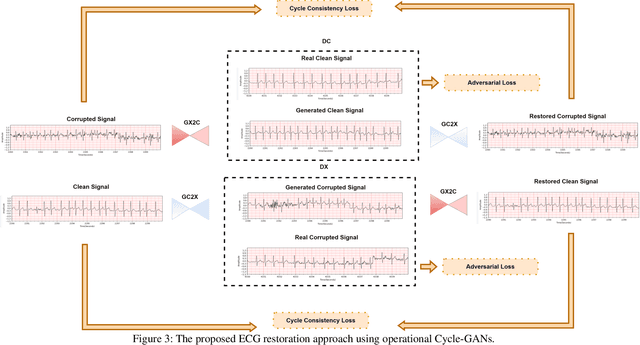
Abstract:Continuous long-term monitoring of electrocardiography (ECG) signals is crucial for the early detection of cardiac abnormalities such as arrhythmia. Non-clinical ECG recordings acquired by Holter and wearable ECG sensors often suffer from severe artifacts such as baseline wander, signal cuts, motion artifacts, variations on QRS amplitude, noise, and other interferences. Usually, a set of such artifacts occur on the same ECG signal with varying severity and duration, and this makes an accurate diagnosis by machines or medical doctors extremely difficult. Despite numerous studies that have attempted ECG denoising, they naturally fail to restore the actual ECG signal corrupted with such artifacts due to their simple and naive noise model. In this study, we propose a novel approach for blind ECG restoration using cycle-consistent generative adversarial networks (Cycle-GANs) where the quality of the signal can be improved to a clinical level ECG regardless of the type and severity of the artifacts corrupting the signal. To further boost the restoration performance, we propose 1D operational Cycle-GANs with the generative neuron model. The proposed approach has been evaluated extensively using one of the largest benchmark ECG datasets from the China Physiological Signal Challenge (CPSC-2020) with more than one million beats. Besides the quantitative and qualitative evaluations, a group of cardiologists performed medical evaluations to validate the quality and usability of the restored ECG, especially for an accurate arrhythmia diagnosis.
A Shallow U-Net Architecture for Reliably Predicting Blood Pressure from Photoplethysmogram and Electrocardiogram Signals
Nov 12, 2021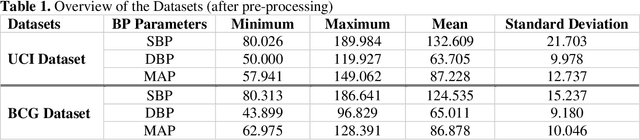
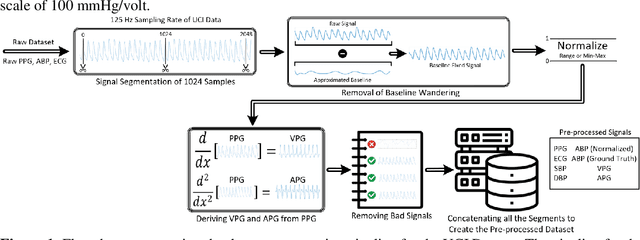
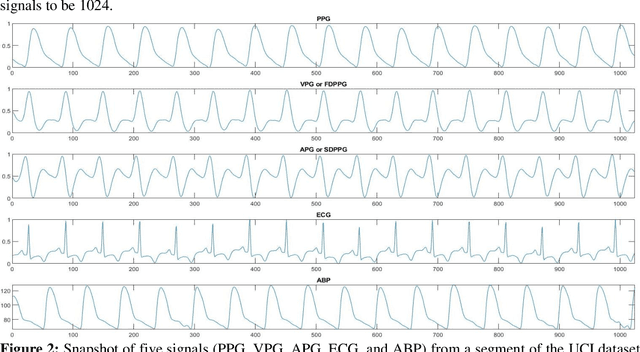
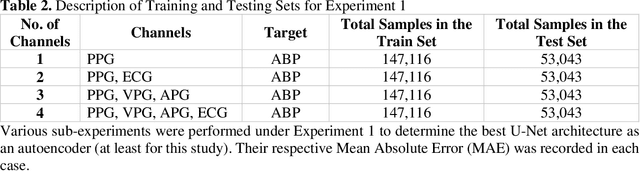
Abstract:Cardiovascular diseases are the most common causes of death around the world. To detect and treat heart-related diseases, continuous Blood Pressure (BP) monitoring along with many other parameters are required. Several invasive and non-invasive methods have been developed for this purpose. Most existing methods used in the hospitals for continuous monitoring of BP are invasive. On the contrary, cuff-based BP monitoring methods, which can predict Systolic Blood Pressure (SBP) and Diastolic Blood Pressure (DBP), cannot be used for continuous monitoring. Several studies attempted to predict BP from non-invasively collectible signals such as Photoplethysmogram (PPG) and Electrocardiogram (ECG), which can be used for continuous monitoring. In this study, we explored the applicability of autoencoders in predicting BP from PPG and ECG signals. The investigation was carried out on 12,000 instances of 942 patients of the MIMIC-II dataset and it was found that a very shallow, one-dimensional autoencoder can extract the relevant features to predict the SBP and DBP with the state-of-the-art performance on a very large dataset. Independent test set from a portion of the MIMIC-II dataset provides an MAE of 2.333 and 0.713 for SBP and DBP, respectively. On an external dataset of forty subjects, the model trained on the MIMIC-II dataset, provides an MAE of 2.728 and 1.166 for SBP and DBP, respectively. For both the cases, the results met British Hypertension Society (BHS) Grade A and surpassed the studies from the current literature.
A Machine Learning Model for Early Detection of Diabetic Foot using Thermogram Images
Jun 27, 2021



Abstract:Diabetes foot ulceration (DFU) and amputation are a cause of significant morbidity. The prevention of DFU may be achieved by the identification of patients at risk of DFU and the institution of preventative measures through education and offloading. Several studies have reported that thermogram images may help to detect an increase in plantar temperature prior to DFU. However, the distribution of plantar temperature may be heterogeneous, making it difficult to quantify and utilize to predict outcomes. We have compared a machine learning-based scoring technique with feature selection and optimization techniques and learning classifiers to several state-of-the-art Convolutional Neural Networks (CNNs) on foot thermogram images and propose a robust solution to identify the diabetic foot. A comparatively shallow CNN model, MobilenetV2 achieved an F1 score of ~95% for a two-feet thermogram image-based classification and the AdaBoost Classifier used 10 features and achieved an F1 score of 97 %. A comparison of the inference time for the best-performing networks confirmed that the proposed algorithm can be deployed as a smartphone application to allow the user to monitor the progression of the DFU in a home setting.
COV-ECGNET: COVID-19 detection using ECG trace images with deep convolutional neural network
Jun 01, 2021



Abstract:The reliable and rapid identification of the COVID-19 has become crucial to prevent the rapid spread of the disease, ease lockdown restrictions and reduce pressure on public health infrastructures. Recently, several methods and techniques have been proposed to detect the SARS-CoV-2 virus using different images and data. However, this is the first study that will explore the possibility of using deep convolutional neural network (CNN) models to detect COVID-19 from electrocardiogram (ECG) trace images. In this work, COVID-19 and other cardiovascular diseases (CVDs) were detected using deep-learning techniques. A public dataset of ECG images consists of 1937 images from five distinct categories, such as Normal, COVID-19, myocardial infarction (MI), abnormal heartbeat (AHB), and recovered myocardial infarction (RMI) were used in this study. Six different deep CNN models (ResNet18, ResNet50, ResNet101, InceptionV3, DenseNet201, and MobileNetv2) were used to investigate three different classification schemes: two-class classification (Normal vs COVID-19); three-class classification (Normal, COVID-19, and Other CVDs), and finally, five-class classification (Normal, COVID-19, MI, AHB, and RMI). For two-class and three-class classification, Densenet201 outperforms other networks with an accuracy of 99.1%, and 97.36%, respectively; while for the five-class classification, InceptionV3 outperforms others with an accuracy of 97.83%. ScoreCAM visualization confirms that the networks are learning from the relevant area of the trace images. Since the proposed method uses ECG trace images which can be captured by smartphones and are readily available facilities in low-resources countries, this study will help in faster computer-aided diagnosis of COVID-19 and other cardiac abnormalities.
QUCoughScope: An Artificially Intelligent Mobile Application to Detect Asymptomatic COVID-19 Patients using Cough and Breathing Sounds
Mar 20, 2021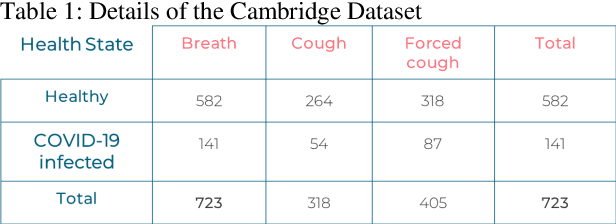
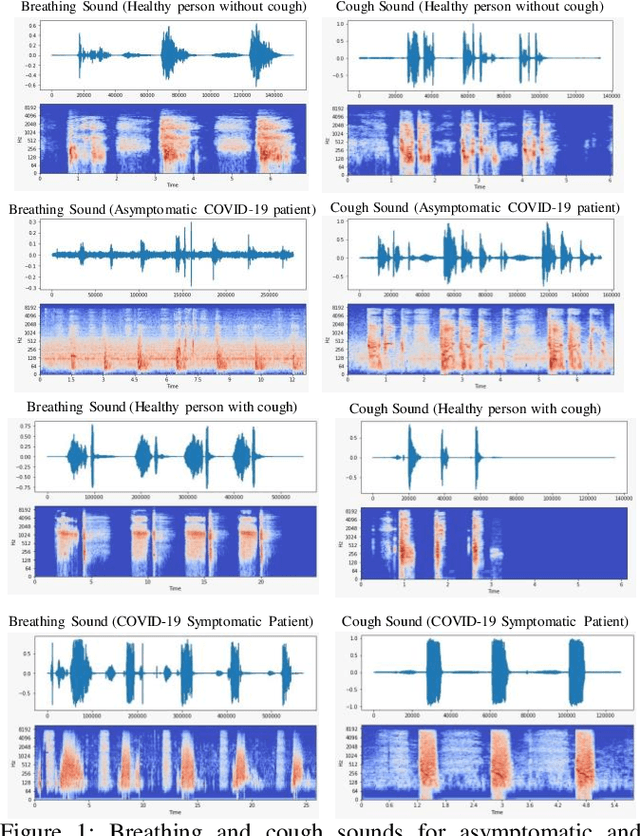
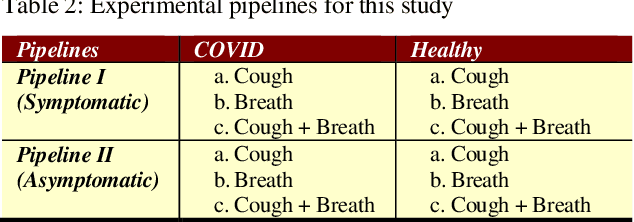
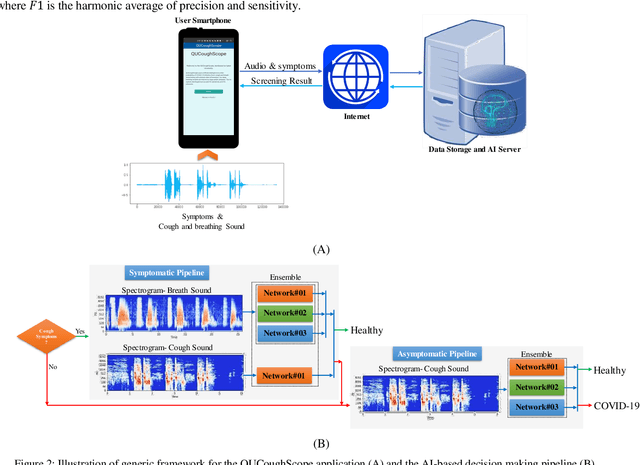
Abstract:In the break of COVID-19 pandemic, mass testing has become essential to reduce the spread of the virus. Several recent studies suggest that a significant number of COVID-19 patients display no physical symptoms whatsoever. Therefore, it is unlikely that these patients will undergo COVID-19 test, which increases their chances of unintentionally spreading the virus. Currently, the primary diagnostic tool to detect COVID-19 is RT-PCR test on collected respiratory specimens from the suspected case. This requires patients to travel to a laboratory facility to be tested, thereby potentially infecting others along the way.It is evident from recent researches that asymptomatic COVID-19 patients cough and breath in a different way than the healthy people. Several research groups have created mobile and web-platform for crowdsourcing the symptoms, cough and breathing sounds from healthy, COVID-19 and Non-COVID patients. Some of these data repositories were made public. We have received such a repository from Cambridge University team under data-sharing agreement, where we have cough and breathing sound samples for 582 and 141 healthy and COVID-19 patients, respectively. 87 COVID-19 patients were asymptomatic, while rest of them have cough. We have developed an Android application to automatically screen COVID-19 from the comfort of people homes. Test subjects can simply download a mobile application, enter their symptoms, record an audio clip of their cough and breath, and upload the data anonymously to our servers. Our backend server converts the audio clip to spectrogram and then apply our state-of-the-art machine learning model to classify between cough sounds produced by COVID-19 patients, as opposed to healthy subjects or those with other respiratory conditions. The system can detect asymptomatic COVID-19 patients with a sensitivity more than 91%.
COVID-19 Infection Localization and Severity Grading from Chest X-ray Images
Mar 14, 2021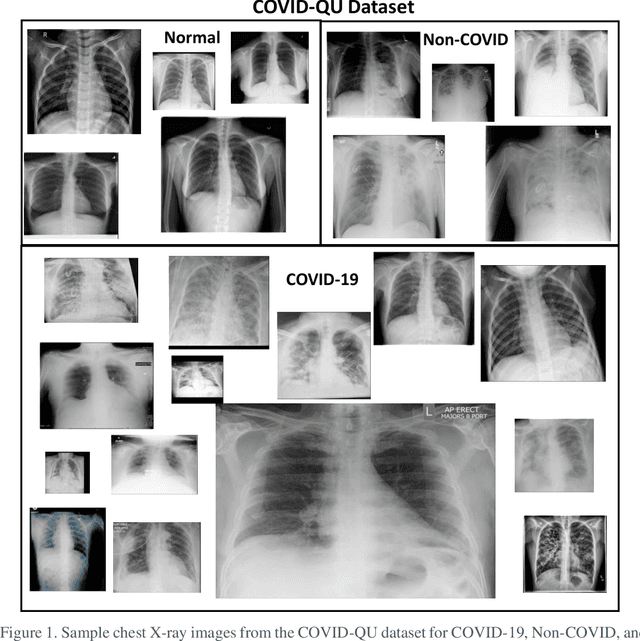
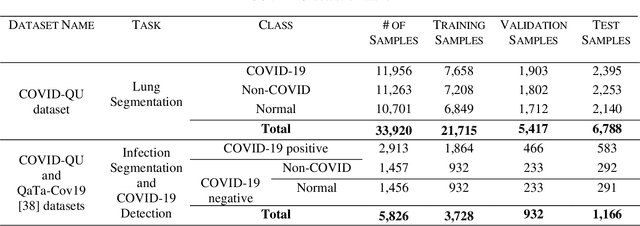
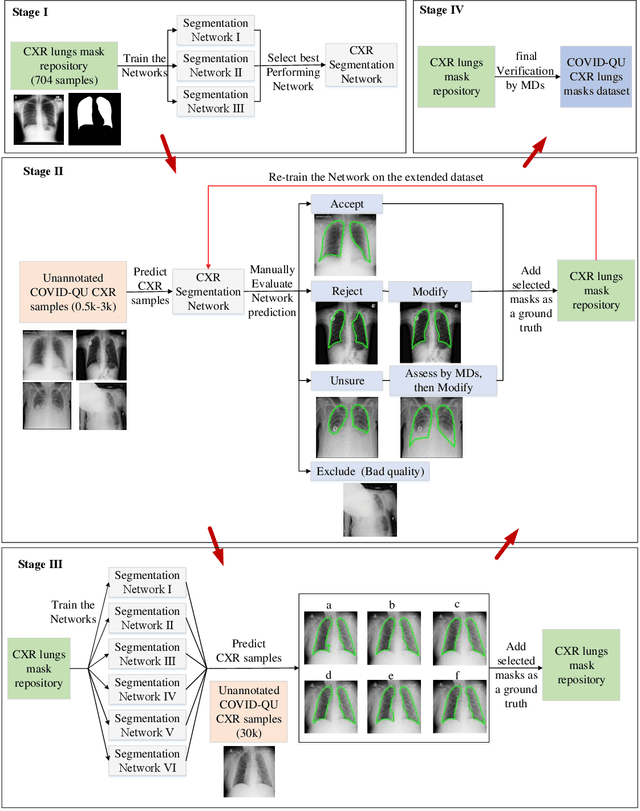
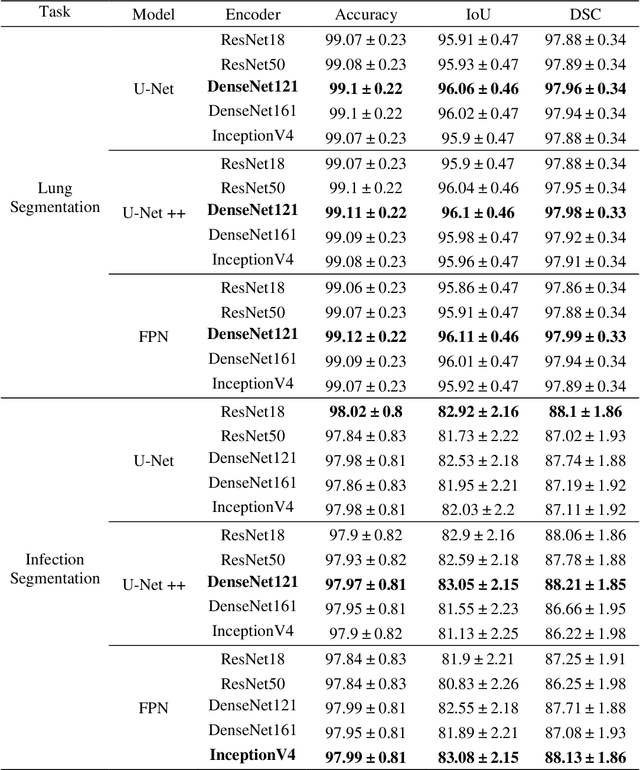
Abstract:Coronavirus disease 2019 (COVID-19) has been the main agenda of the whole world, since it came into sight in December 2019 as it has significantly affected the world economy and healthcare system. Given the effects of COVID-19 on pulmonary tissues, chest radiographic imaging has become a necessity for screening and monitoring the disease. Numerous studies have proposed Deep Learning approaches for the automatic diagnosis of COVID-19. Although these methods achieved astonishing performance in detection, they have used limited chest X-ray (CXR) repositories for evaluation, usually with a few hundred COVID-19 CXR images only. Thus, such data scarcity prevents reliable evaluation with the potential of overfitting. In addition, most studies showed no or limited capability in infection localization and severity grading of COVID-19 pneumonia. In this study, we address this urgent need by proposing a systematic and unified approach for lung segmentation and COVID-19 localization with infection quantification from CXR images. To accomplish this, we have constructed the largest benchmark dataset with 33,920 CXR images, including 11,956 COVID-19 samples, where the annotation of ground-truth lung segmentation masks is performed on CXRs by a novel human-machine collaborative approach. An extensive set of experiments was performed using the state-of-the-art segmentation networks, U-Net, U-Net++, and Feature Pyramid Networks (FPN). The developed network, after an extensive iterative process, reached a superior performance for lung region segmentation with Intersection over Union (IoU) of 96.11% and Dice Similarity Coefficient (DSC) of 97.99%. Furthermore, COVID-19 infections of various shapes and types were reliably localized with 83.05% IoU and 88.21% DSC. Finally, the proposed approach has achieved an outstanding COVID-19 detection performance with both sensitivity and specificity values above 99%.
EDITH :ECG biometrics aided by Deep learning for reliable Individual auTHentication
Feb 16, 2021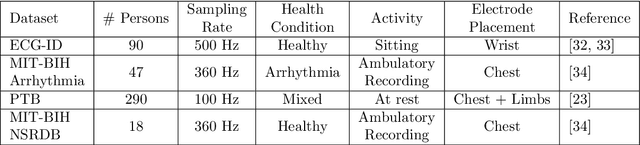
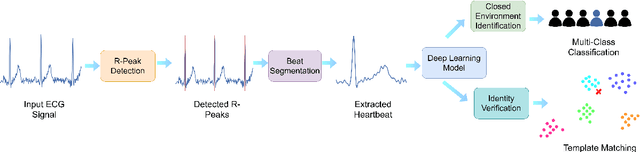
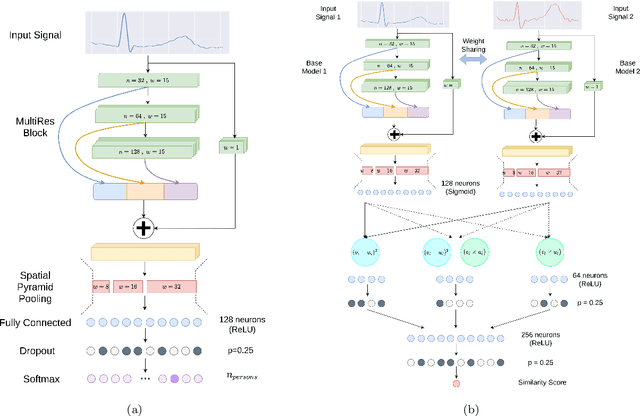
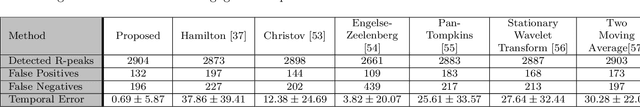
Abstract:In recent years, physiological signal based authentication has shown great promises,for its inherent robustness against forgery. Electrocardiogram (ECG) signal, being the most widely studied biosignal, has also received the highest level of attention in this regard. It has been proven with numerous studies that by analyzing ECG signals from different persons, it is possible to identify them, with acceptable accuracy. In this work, we present, EDITH, a deep learning-based framework for ECG biometrics authentication system. Moreover, we hypothesize and demonstrate that Siamese architectures can be used over typical distance metrics for improved performance. We have evaluated EDITH using 4 commonly used datasets and outperformed the prior works using less number of beats. EDITH performs competitively using just a single heartbeat (96-99.75% accuracy) and can be further enhanced by fusing multiple beats (100% accuracy from 3 to 6 beats). Furthermore, the proposed Siamese architecture manages to reduce the identity verification Equal Error Rate (EER) to 1.29%. A limited case study of EDITH with real-world experimental data also suggests its potential as a practical authentication system.
Detection and severity classification of COVID-19 in CT images using deep learning
Feb 15, 2021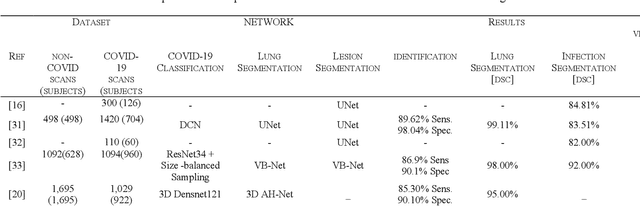
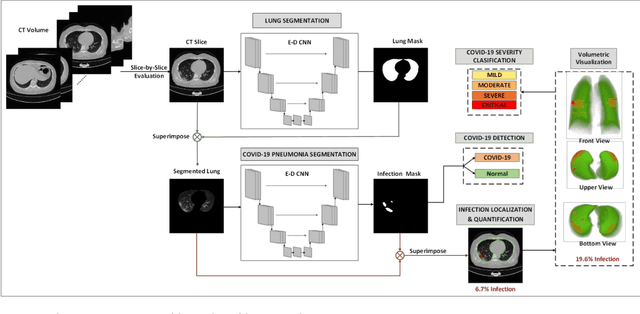
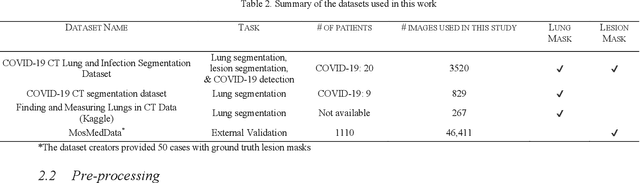
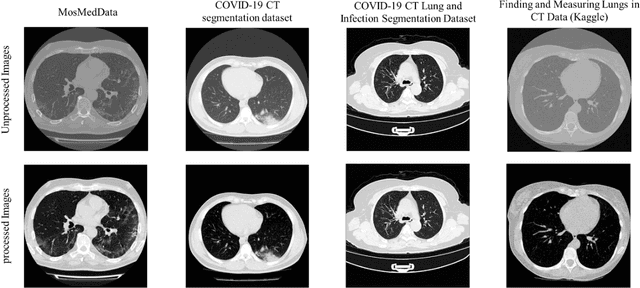
Abstract:Since the breakout of coronavirus disease (COVID-19), the computer-aided diagnosis has become a necessity to prevent the spread of the virus. Detecting COVID-19 at an early stage is essential to reduce the mortality risk of the patients. In this study, a cascaded system is proposed to segment the lung, detect, localize, and quantify COVID-19 infections from computed tomography (CT) images Furthermore, the system classifies the severity of COVID-19 as mild, moderate, severe, or critical based on the percentage of infected lungs. An extensive set of experiments were performed using state-of-the-art deep Encoder-Decoder Convolutional Neural Networks (ED-CNNs), UNet, and Feature Pyramid Network (FPN), with different backbone (encoder) structures using the variants of DenseNet and ResNet. The conducted experiments showed the best performance for lung region segmentation with Dice Similarity Coefficient (DSC) of 97.19% and Intersection over Union (IoU) of 95.10% using U-Net model with the DenseNet 161 encoder. Furthermore, the proposed system achieved an elegant performance for COVID-19 infection segmentation with a DSC of 94.13% and IoU of 91.85% using the FPN model with the DenseNet201 encoder. The achieved performance is significantly superior to previous methods for COVID-19 lesion localization. Besides, the proposed system can reliably localize infection of various shapes and sizes, especially small infection regions, which are rarely considered in recent studies. Moreover, the proposed system achieved high COVID-19 detection performance with 99.64% sensitivity and 98.72% specificity. Finally, the system was able to discriminate between different severity levels of COVID-19 infection over a dataset of 1,110 subjects with sensitivity values of 98.3%, 71.2%, 77.8%, and 100% for mild, moderate, severe, and critical infections, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge