Sakib Mahmud
CASR-Net: An Image Processing-focused Deep Learning-based Coronary Artery Segmentation and Refinement Network for X-ray Coronary Angiogram
Oct 31, 2025Abstract:Early detection of coronary artery disease (CAD) is critical for reducing mortality and improving patient treatment planning. While angiographic image analysis from X-rays is a common and cost-effective method for identifying cardiac abnormalities, including stenotic coronary arteries, poor image quality can significantly impede clinical diagnosis. We present the Coronary Artery Segmentation and Refinement Network (CASR-Net), a three-stage pipeline comprising image preprocessing, segmentation, and refinement. A novel multichannel preprocessing strategy combining CLAHE and an improved Ben Graham method provides incremental gains, increasing Dice Score Coefficient (DSC) by 0.31-0.89% and Intersection over Union (IoU) by 0.40-1.16% compared with using the techniques individually. The core innovation is a segmentation network built on a UNet with a DenseNet121 encoder and a Self-organized Operational Neural Network (Self-ONN) based decoder, which preserves the continuity of narrow and stenotic vessel branches. A final contour refinement module further suppresses false positives. Evaluated with 5-fold cross-validation on a combination of two public datasets that contain both healthy and stenotic arteries, CASR-Net outperformed several state-of-the-art models, achieving an IoU of 61.43%, a DSC of 76.10%, and clDice of 79.36%. These results highlight a robust approach to automated coronary artery segmentation, offering a valuable tool to support clinicians in diagnosis and treatment planning.
A Shallow U-Net Architecture for Reliably Predicting Blood Pressure from Photoplethysmogram and Electrocardiogram Signals
Nov 12, 2021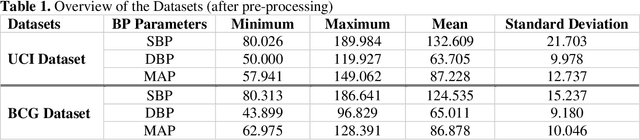
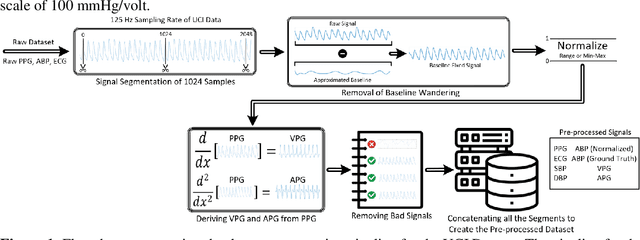
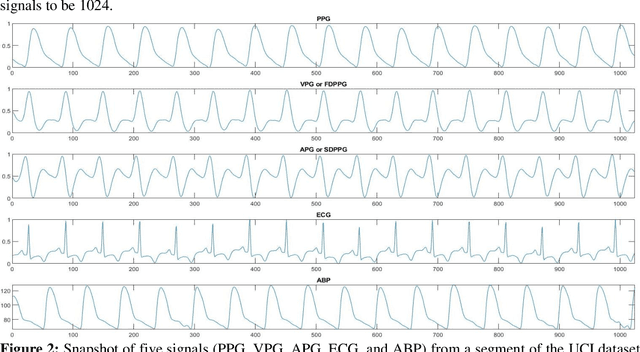
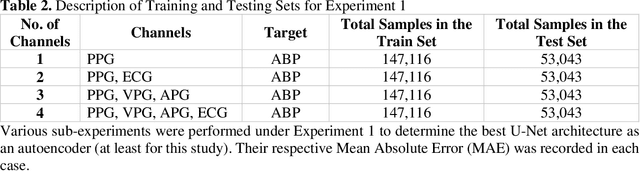
Abstract:Cardiovascular diseases are the most common causes of death around the world. To detect and treat heart-related diseases, continuous Blood Pressure (BP) monitoring along with many other parameters are required. Several invasive and non-invasive methods have been developed for this purpose. Most existing methods used in the hospitals for continuous monitoring of BP are invasive. On the contrary, cuff-based BP monitoring methods, which can predict Systolic Blood Pressure (SBP) and Diastolic Blood Pressure (DBP), cannot be used for continuous monitoring. Several studies attempted to predict BP from non-invasively collectible signals such as Photoplethysmogram (PPG) and Electrocardiogram (ECG), which can be used for continuous monitoring. In this study, we explored the applicability of autoencoders in predicting BP from PPG and ECG signals. The investigation was carried out on 12,000 instances of 942 patients of the MIMIC-II dataset and it was found that a very shallow, one-dimensional autoencoder can extract the relevant features to predict the SBP and DBP with the state-of-the-art performance on a very large dataset. Independent test set from a portion of the MIMIC-II dataset provides an MAE of 2.333 and 0.713 for SBP and DBP, respectively. On an external dataset of forty subjects, the model trained on the MIMIC-II dataset, provides an MAE of 2.728 and 1.166 for SBP and DBP, respectively. For both the cases, the results met British Hypertension Society (BHS) Grade A and surpassed the studies from the current literature.
QUCoughScope: An Artificially Intelligent Mobile Application to Detect Asymptomatic COVID-19 Patients using Cough and Breathing Sounds
Mar 20, 2021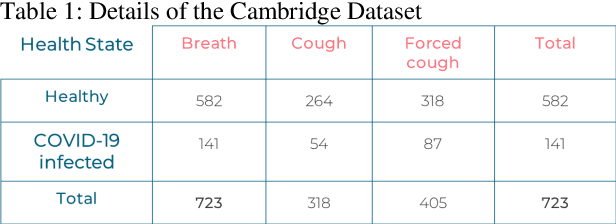
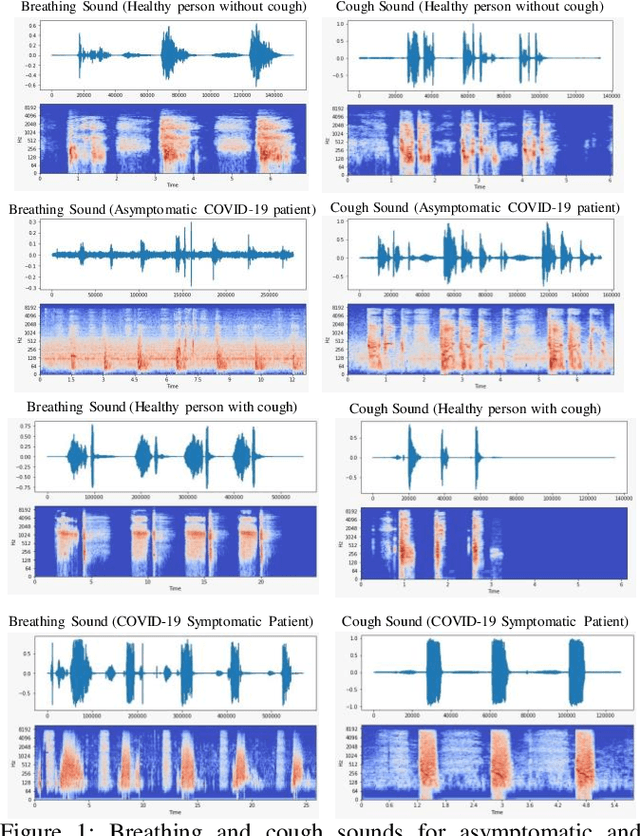
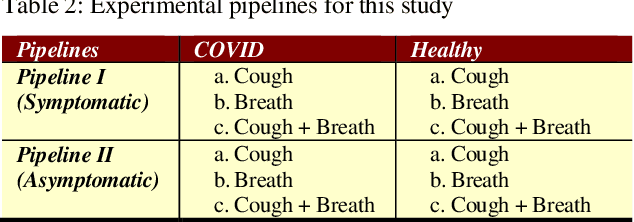
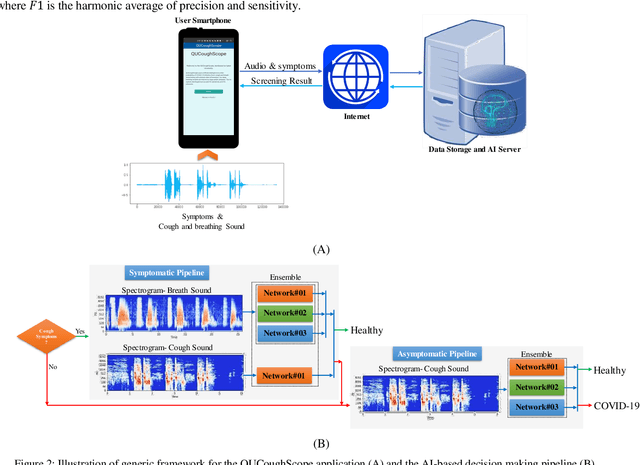
Abstract:In the break of COVID-19 pandemic, mass testing has become essential to reduce the spread of the virus. Several recent studies suggest that a significant number of COVID-19 patients display no physical symptoms whatsoever. Therefore, it is unlikely that these patients will undergo COVID-19 test, which increases their chances of unintentionally spreading the virus. Currently, the primary diagnostic tool to detect COVID-19 is RT-PCR test on collected respiratory specimens from the suspected case. This requires patients to travel to a laboratory facility to be tested, thereby potentially infecting others along the way.It is evident from recent researches that asymptomatic COVID-19 patients cough and breath in a different way than the healthy people. Several research groups have created mobile and web-platform for crowdsourcing the symptoms, cough and breathing sounds from healthy, COVID-19 and Non-COVID patients. Some of these data repositories were made public. We have received such a repository from Cambridge University team under data-sharing agreement, where we have cough and breathing sound samples for 582 and 141 healthy and COVID-19 patients, respectively. 87 COVID-19 patients were asymptomatic, while rest of them have cough. We have developed an Android application to automatically screen COVID-19 from the comfort of people homes. Test subjects can simply download a mobile application, enter their symptoms, record an audio clip of their cough and breath, and upload the data anonymously to our servers. Our backend server converts the audio clip to spectrogram and then apply our state-of-the-art machine learning model to classify between cough sounds produced by COVID-19 patients, as opposed to healthy subjects or those with other respiratory conditions. The system can detect asymptomatic COVID-19 patients with a sensitivity more than 91%.
COVID-19 Infection Localization and Severity Grading from Chest X-ray Images
Mar 14, 2021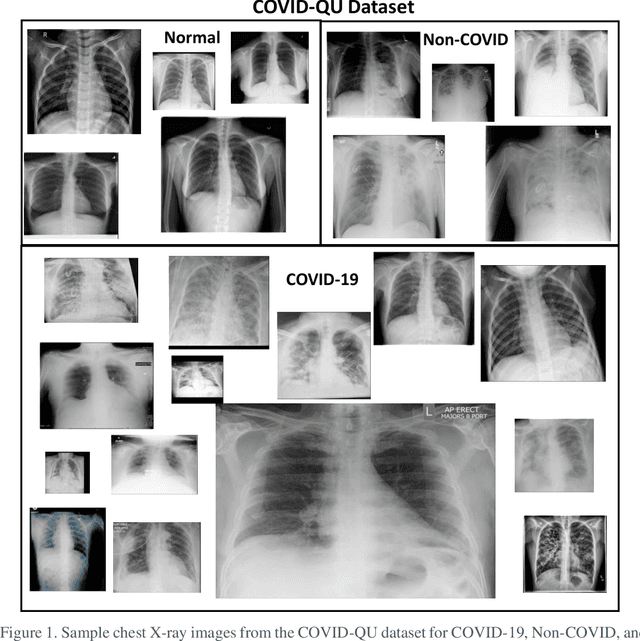
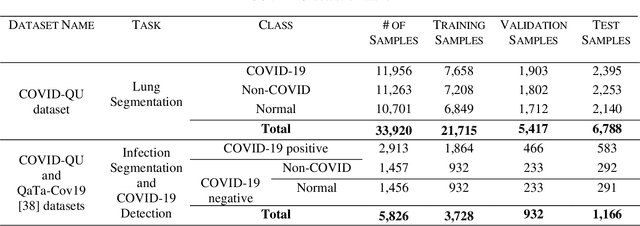
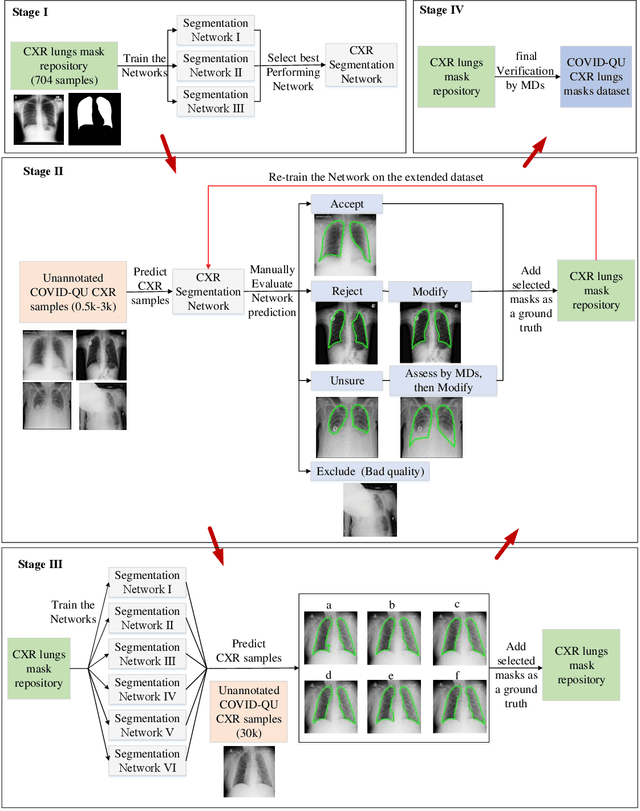
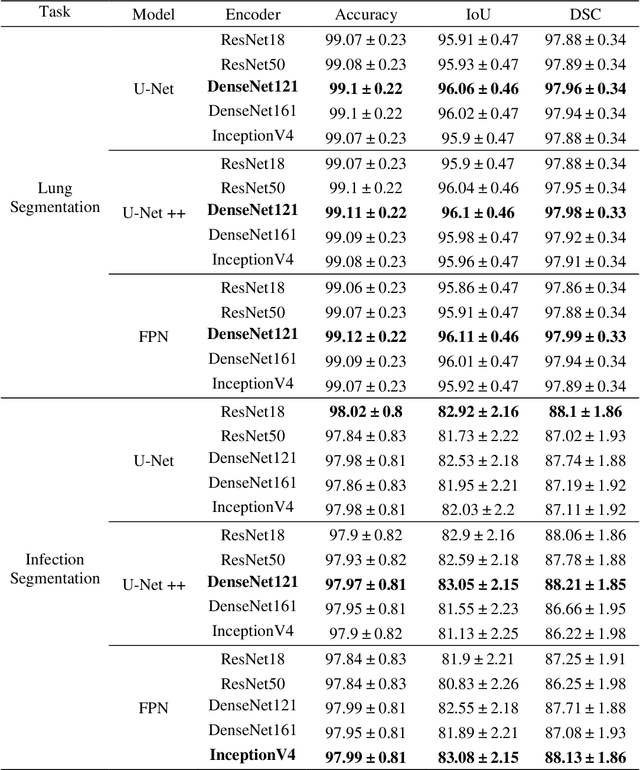
Abstract:Coronavirus disease 2019 (COVID-19) has been the main agenda of the whole world, since it came into sight in December 2019 as it has significantly affected the world economy and healthcare system. Given the effects of COVID-19 on pulmonary tissues, chest radiographic imaging has become a necessity for screening and monitoring the disease. Numerous studies have proposed Deep Learning approaches for the automatic diagnosis of COVID-19. Although these methods achieved astonishing performance in detection, they have used limited chest X-ray (CXR) repositories for evaluation, usually with a few hundred COVID-19 CXR images only. Thus, such data scarcity prevents reliable evaluation with the potential of overfitting. In addition, most studies showed no or limited capability in infection localization and severity grading of COVID-19 pneumonia. In this study, we address this urgent need by proposing a systematic and unified approach for lung segmentation and COVID-19 localization with infection quantification from CXR images. To accomplish this, we have constructed the largest benchmark dataset with 33,920 CXR images, including 11,956 COVID-19 samples, where the annotation of ground-truth lung segmentation masks is performed on CXRs by a novel human-machine collaborative approach. An extensive set of experiments was performed using the state-of-the-art segmentation networks, U-Net, U-Net++, and Feature Pyramid Networks (FPN). The developed network, after an extensive iterative process, reached a superior performance for lung region segmentation with Intersection over Union (IoU) of 96.11% and Dice Similarity Coefficient (DSC) of 97.99%. Furthermore, COVID-19 infections of various shapes and types were reliably localized with 83.05% IoU and 88.21% DSC. Finally, the proposed approach has achieved an outstanding COVID-19 detection performance with both sensitivity and specificity values above 99%.
Detection and severity classification of COVID-19 in CT images using deep learning
Feb 15, 2021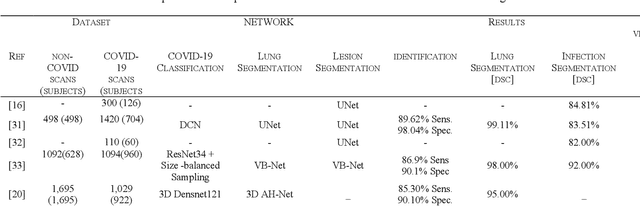
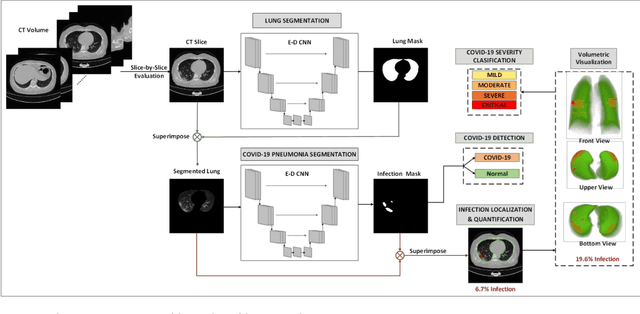
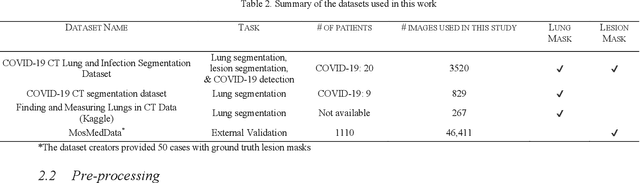
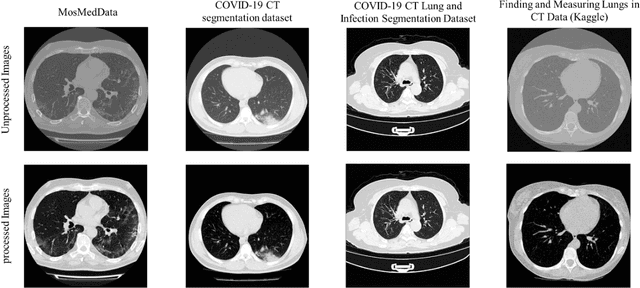
Abstract:Since the breakout of coronavirus disease (COVID-19), the computer-aided diagnosis has become a necessity to prevent the spread of the virus. Detecting COVID-19 at an early stage is essential to reduce the mortality risk of the patients. In this study, a cascaded system is proposed to segment the lung, detect, localize, and quantify COVID-19 infections from computed tomography (CT) images Furthermore, the system classifies the severity of COVID-19 as mild, moderate, severe, or critical based on the percentage of infected lungs. An extensive set of experiments were performed using state-of-the-art deep Encoder-Decoder Convolutional Neural Networks (ED-CNNs), UNet, and Feature Pyramid Network (FPN), with different backbone (encoder) structures using the variants of DenseNet and ResNet. The conducted experiments showed the best performance for lung region segmentation with Dice Similarity Coefficient (DSC) of 97.19% and Intersection over Union (IoU) of 95.10% using U-Net model with the DenseNet 161 encoder. Furthermore, the proposed system achieved an elegant performance for COVID-19 infection segmentation with a DSC of 94.13% and IoU of 91.85% using the FPN model with the DenseNet201 encoder. The achieved performance is significantly superior to previous methods for COVID-19 lesion localization. Besides, the proposed system can reliably localize infection of various shapes and sizes, especially small infection regions, which are rarely considered in recent studies. Moreover, the proposed system achieved high COVID-19 detection performance with 99.64% sensitivity and 98.72% specificity. Finally, the system was able to discriminate between different severity levels of COVID-19 infection over a dataset of 1,110 subjects with sensitivity values of 98.3%, 71.2%, 77.8%, and 100% for mild, moderate, severe, and critical infections, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge