Tahir Hamid
Echocardiography to Cardiac MRI View Transformation for Real-Time Blind Restoration
Dec 09, 2024



Abstract:Echocardiography is the most widely used imaging to monitor cardiac functions, serving as the first line in early detection of myocardial ischemia and infarction. However, echocardiography often suffers from several artifacts including sensor noise, lack of contrast, severe saturation, and missing myocardial segments which severely limit its usage in clinical diagnosis. In recent years, several machine learning methods have been proposed to improve echocardiography views. Yet, these methods usually address only a specific problem (e.g. denoising) and thus cannot provide a robust and reliable restoration in general. On the other hand, cardiac MRI provides a clean view of the heart without suffering such severe issues. However, due to its significantly higher cost, it is often only afforded by a few major hospitals, hence hindering its use and accessibility. In this pilot study, we propose a novel approach to transform echocardiography into the cardiac MRI view. For this purpose, Echo2MRI dataset, consisting of echocardiography and real cardiac MRI image pairs, is composed and will be shared publicly. A dedicated Cycle-consistent Generative Adversarial Network (Cycle-GAN) is trained to learn the transformation from echocardiography frames to cardiac MRI views. An extensive set of qualitative evaluations shows that the proposed transformer can synthesize high-quality artifact-free synthetic cardiac MRI views from a given sequence of echocardiography frames. Medical evaluations performed by a group of cardiologists further demonstrate that synthetic MRI views are indistinguishable from their original counterparts and are preferred over their initial sequence of echocardiography frames for diagnosis in 78.9% of the cases.
R2C-GAN: Restore-to-Classify GANs for Blind X-Ray Restoration and COVID-19 Classification
Sep 29, 2022



Abstract:Restoration of poor quality images with a blended set of artifacts plays a vital role for a reliable diagnosis. Existing studies have focused on specific restoration problems such as image deblurring, denoising, and exposure correction where there is usually a strong assumption on the artifact type and severity. As a pioneer study in blind X-ray restoration, we propose a joint model for generic image restoration and classification: Restore-to-Classify Generative Adversarial Networks (R2C-GANs). Such a jointly optimized model keeps any disease intact after the restoration. Therefore, this will naturally lead to a higher diagnosis performance thanks to the improved X-ray image quality. To accomplish this crucial objective, we define the restoration task as an Image-to-Image translation problem from poor quality having noisy, blurry, or over/under-exposed images to high quality image domain. The proposed R2C-GAN model is able to learn forward and inverse transforms between the two domains using unpaired training samples. Simultaneously, the joint classification preserves the disease label during restoration. Moreover, the R2C-GANs are equipped with operational layers/neurons reducing the network depth and further boosting both restoration and classification performances. The proposed joint model is extensively evaluated over the QaTa-COV19 dataset for Coronavirus Disease 2019 (COVID-19) classification. The proposed restoration approach achieves over 90% F1-Score which is significantly higher than the performance of any deep model. Moreover, in the qualitative analysis, the restoration performance of R2C-GANs is approved by a group of medical doctors. We share the software implementation at https://github.com/meteahishali/R2C-GAN.
Blind ECG Restoration by Operational Cycle-GANs
Jan 29, 2022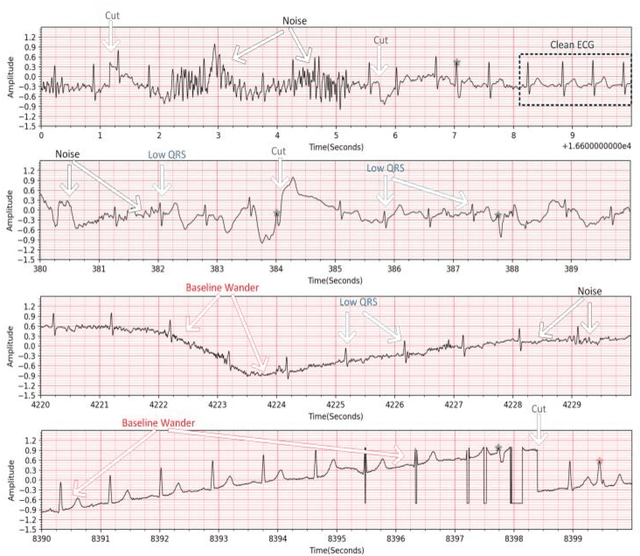
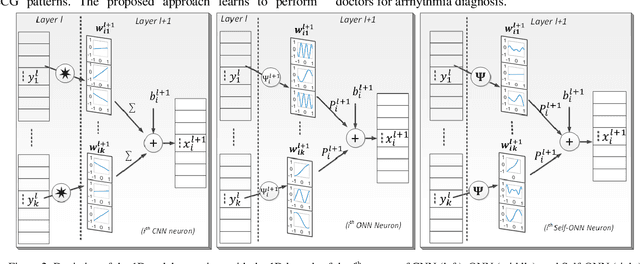
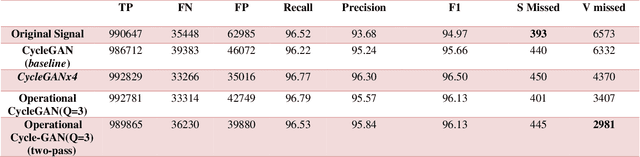
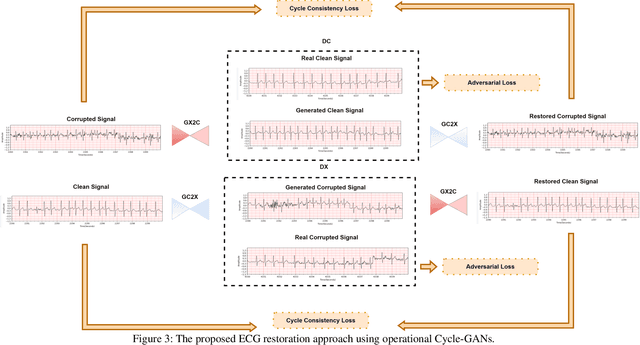
Abstract:Continuous long-term monitoring of electrocardiography (ECG) signals is crucial for the early detection of cardiac abnormalities such as arrhythmia. Non-clinical ECG recordings acquired by Holter and wearable ECG sensors often suffer from severe artifacts such as baseline wander, signal cuts, motion artifacts, variations on QRS amplitude, noise, and other interferences. Usually, a set of such artifacts occur on the same ECG signal with varying severity and duration, and this makes an accurate diagnosis by machines or medical doctors extremely difficult. Despite numerous studies that have attempted ECG denoising, they naturally fail to restore the actual ECG signal corrupted with such artifacts due to their simple and naive noise model. In this study, we propose a novel approach for blind ECG restoration using cycle-consistent generative adversarial networks (Cycle-GANs) where the quality of the signal can be improved to a clinical level ECG regardless of the type and severity of the artifacts corrupting the signal. To further boost the restoration performance, we propose 1D operational Cycle-GANs with the generative neuron model. The proposed approach has been evaluated extensively using one of the largest benchmark ECG datasets from the China Physiological Signal Challenge (CPSC-2020) with more than one million beats. Besides the quantitative and qualitative evaluations, a group of cardiologists performed medical evaluations to validate the quality and usability of the restored ECG, especially for an accurate arrhythmia diagnosis.
Early Myocardial Infarction Detection over Multi-view Echocardiography
Nov 09, 2021



Abstract:Myocardial infarction (MI) is the leading cause of mortality in the world that occurs due to a blockage of the coronary arteries feeding the myocardium. An early diagnosis of MI and its localization can mitigate the extent of myocardial damage by facilitating early therapeutic interventions. Following the blockage of a coronary artery, the regional wall motion abnormality (RWMA) of the ischemic myocardial segments is the earliest change to set in. Echocardiography is the fundamental tool to assess any RWMA. Assessing the motion of the left ventricle (LV) wall only from a single echocardiography view may lead to missing the diagnosis of MI as the RWMA may not be visible on that specific view. Therefore, in this study, we propose to fuse apical 4-chamber (A4C) and apical 2-chamber (A2C) views in which a total of 11 myocardial segments can be analyzed for MI detection. The proposed method first estimates the motion of the LV wall by Active Polynomials (APs), which extract and track the endocardial boundary to compute myocardial segment displacements. The features are extracted from the A4C and A2C view displacements, which are fused and fed into the classifiers to detect MI. The main contributions of this study are 1) creation of a new benchmark dataset by including both A4C and A2C views in a total of 260 echocardiography recordings, which is publicly shared with the research community, 2) improving the performance of the prior work of threshold-based APs by a Machine Learning based approach, and 3) a pioneer MI detection approach via multi-view echocardiography by fusing the information of A4C and A2C views. Experimental results show that the proposed method achieves 90.91% sensitivity and 86.36% precision for MI detection over multi-view echocardiography.
Fully Automated 2D and 3D Convolutional Neural Networks Pipeline for Video Segmentation and Myocardial Infarction Detection in Echocardiography
Mar 26, 2021



Abstract:Cardiac imaging known as echocardiography is a non-invasive tool utilized to produce data including images and videos, which cardiologists use to diagnose cardiac abnormalities in general and myocardial infarction (MI) in particular. Echocardiography machines can deliver abundant amounts of data that need to be quickly analyzed by cardiologists to help them make a diagnosis and treat cardiac conditions. However, the acquired data quality varies depending on the acquisition conditions and the patient's responsiveness to the setup instructions. These constraints are challenging to doctors especially when patients are facing MI and their lives are at stake. In this paper, we propose an innovative real-time end-to-end fully automated model based on convolutional neural networks (CNN) to detect MI depending on regional wall motion abnormalities (RWMA) of the left ventricle (LV) from videos produced by echocardiography. Our model is implemented as a pipeline consisting of a 2D CNN that performs data preprocessing by segmenting the LV chamber from the apical four-chamber (A4C) view, followed by a 3D CNN that performs a binary classification to detect if the segmented echocardiography shows signs of MI. We trained both CNNs on a dataset composed of 165 echocardiography videos each acquired from a distinct patient. The 2D CNN achieved an accuracy of 97.18% on data segmentation while the 3D CNN achieved 90.9% of accuracy, 100% of precision and 95% of recall on MI detection. Our results demonstrate that creating a fully automated system for MI detection is feasible and propitious.
COVID-19 Infection Localization and Severity Grading from Chest X-ray Images
Mar 14, 2021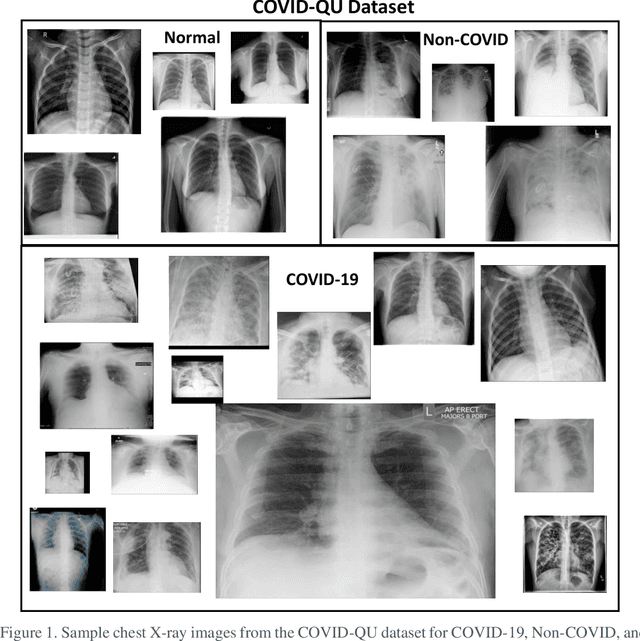
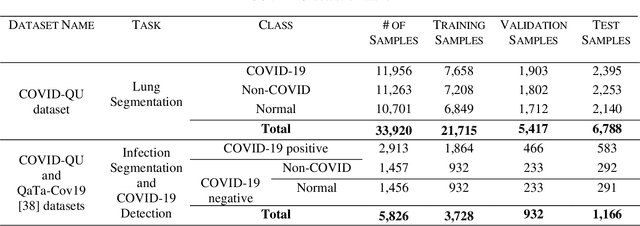
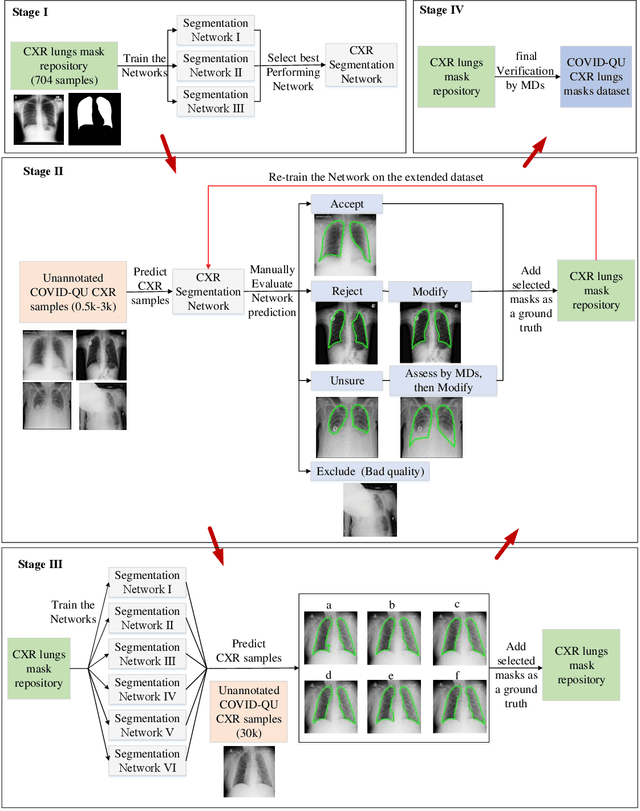
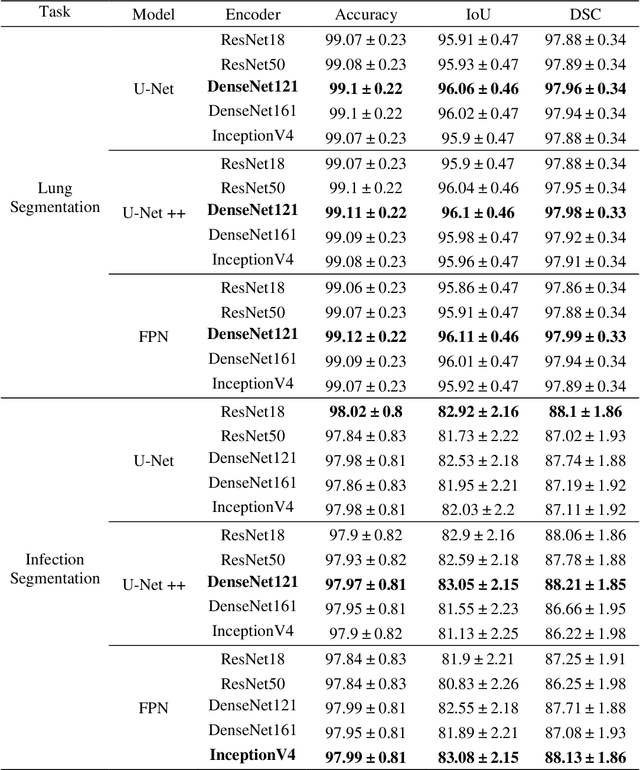
Abstract:Coronavirus disease 2019 (COVID-19) has been the main agenda of the whole world, since it came into sight in December 2019 as it has significantly affected the world economy and healthcare system. Given the effects of COVID-19 on pulmonary tissues, chest radiographic imaging has become a necessity for screening and monitoring the disease. Numerous studies have proposed Deep Learning approaches for the automatic diagnosis of COVID-19. Although these methods achieved astonishing performance in detection, they have used limited chest X-ray (CXR) repositories for evaluation, usually with a few hundred COVID-19 CXR images only. Thus, such data scarcity prevents reliable evaluation with the potential of overfitting. In addition, most studies showed no or limited capability in infection localization and severity grading of COVID-19 pneumonia. In this study, we address this urgent need by proposing a systematic and unified approach for lung segmentation and COVID-19 localization with infection quantification from CXR images. To accomplish this, we have constructed the largest benchmark dataset with 33,920 CXR images, including 11,956 COVID-19 samples, where the annotation of ground-truth lung segmentation masks is performed on CXRs by a novel human-machine collaborative approach. An extensive set of experiments was performed using the state-of-the-art segmentation networks, U-Net, U-Net++, and Feature Pyramid Networks (FPN). The developed network, after an extensive iterative process, reached a superior performance for lung region segmentation with Intersection over Union (IoU) of 96.11% and Dice Similarity Coefficient (DSC) of 97.99%. Furthermore, COVID-19 infections of various shapes and types were reliably localized with 83.05% IoU and 88.21% DSC. Finally, the proposed approach has achieved an outstanding COVID-19 detection performance with both sensitivity and specificity values above 99%.
Early Detection of Myocardial Infarction in Low-Quality Echocardiography
Oct 05, 2020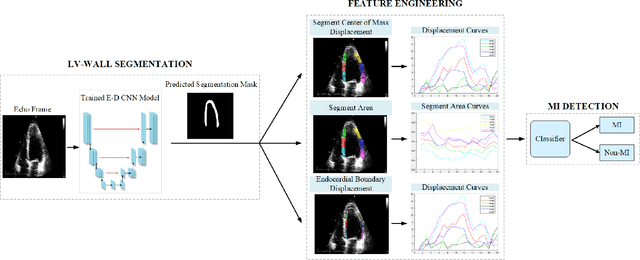

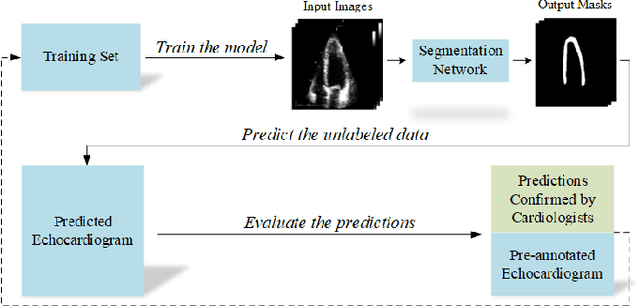
Abstract:Myocardial infarction (MI), or commonly known as heart attack, is a life-threatening worldwide health problem from which 32.4 million of people suffer each year. Early diagnosis and treatment of MI are crucial to prevent further heart tissue damages. However, MI detection in early stages is challenging because the symptoms are not easy to distinguish in electrocardiography findings or biochemical marker values found in the blood. Echocardiography is a noninvasive clinical tool for a more accurate early MI diagnosis, which is used to analyze the regional wall motion abnormalities. When echocardiography quality is poor, the diagnosis becomes a challenging and sometimes infeasible task even for a cardiologist. In this paper, we introduce a three-phase approach for early MI detection in low-quality echocardiography: 1) segmentation of the entire left ventricle (LV) wall of the heart using state-of-the-art deep learning model, 2) analysis of the segmented LV wall by feature engineering, and 3) early MI detection. The main contributions of this study are: highly accurate segmentation of the LV wall from low-resolution (both temporal and spatial) and noisy echocardiographic data, generating the segmentation ground-truth at pixel-level for the unannotated dataset using pseudo labeling approach, and composition of the first public echocardiographic dataset (HMC-QU) labeled by the cardiologists at the Hamad Medical Corporation Hospital in Qatar. Furthermore, the outputs of the proposed approach can significantly help cardiologists for a better assessment of the LV wall characteristics. The proposed method is evaluated in a 5-fold cross validation scheme on the HMC-QU dataset. The proposed approach has achieved an average level of 95.72% sensitivity and 99.58% specificity for the LV wall segmentation, and 85.97% sensitivity, 74.03% specificity, and 86.85% precision for MI detection.
COVID-19 Infection Map Generation and Detection from Chest X-Ray Images
Sep 26, 2020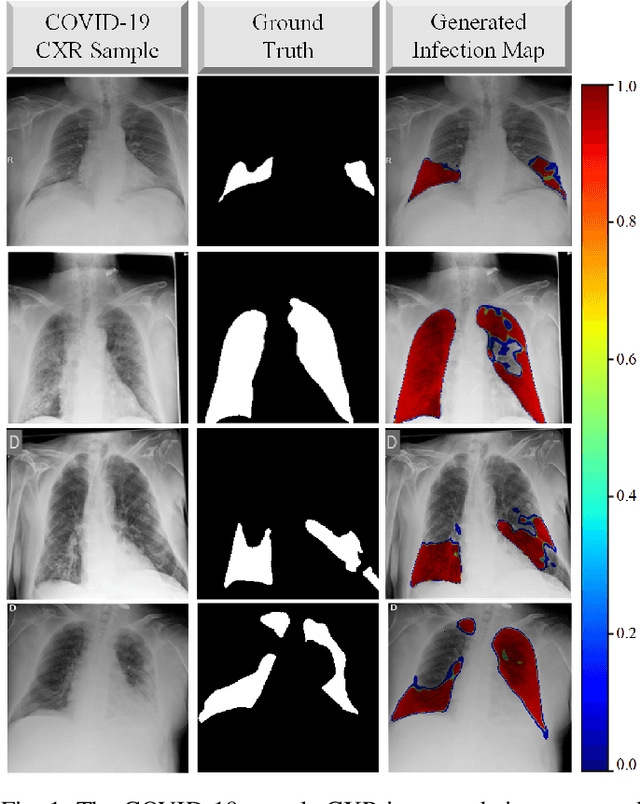
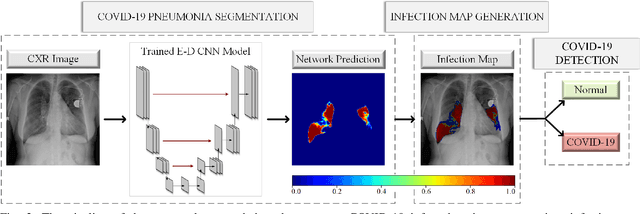
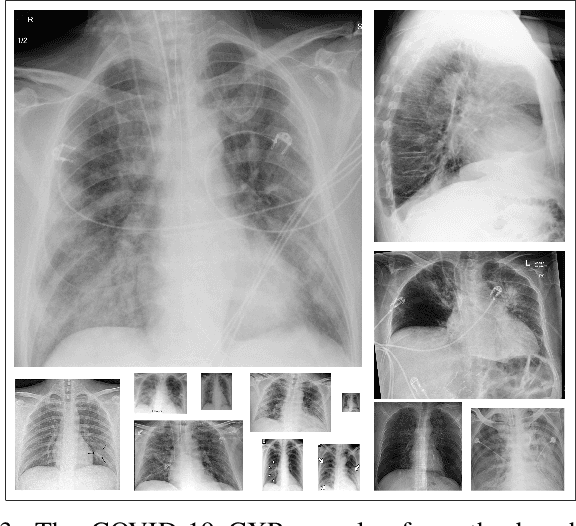
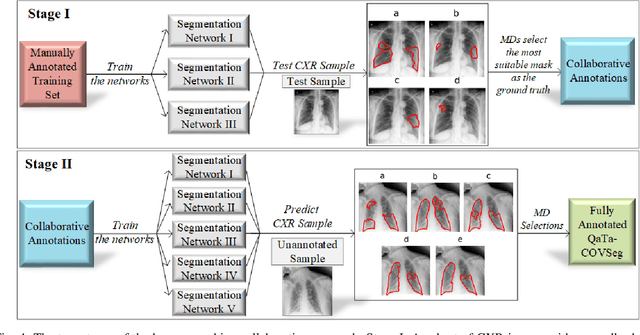
Abstract:Computer-aided diagnosis has become a necessity for accurate and immediate coronavirus disease 2019 (COVID-19) detection to aid treatment and prevent the spread of the virus. Compared to other diagnosis methodologies, chest X-ray (CXR) imaging is an advantageous tool since it is fast, low-cost, and easily accessible. Thus, CXR has a great potential not only to help diagnose COVID-19 but also to track the progression of the disease. Numerous studies have proposed to use Deep Learning techniques for COVID-19 diagnosis. However, they have used very limited CXR image repositories for evaluation with a small number, a few hundreds, of COVID-19 samples. Moreover, these methods can neither localize nor grade the severity of COVID-19 infection. For this purpose, recent studies proposed to explore the activation maps of deep networks. However, they remain inaccurate for localizing the actual infestation making them unreliable for clinical use. This study proposes a novel method for the joint localization, severity grading, and detection of COVID-19 from CXR images by generating the so-called infection maps that can accurately localize and grade the severity of COVID-19 infection. To accomplish this, we have compiled the largest COVID-19 dataset up to date with 2951 COVID-19 CXR images, where the annotation of the ground-truth segmentation masks is performed on CXRs by a novel collaborative expert human-machine approach. Furthermore, we publicly release the first CXR dataset with the ground-truth segmentation masks of the COVID-19 infected regions. A detailed set of experiments show that state-of-the-art segmentation networks can learn to localize COVID-19 infection with an F1-score of 85.81%, that is significantly superior to the activation maps created by the previous methods. Finally, the proposed approach achieved a COVID-19 detection performance with 98.37% sensitivity and 99.16% specificity.
Left Ventricular Wall Motion Estimation by Active Polynomials for Acute Myocardial Infarction Detection
Aug 11, 2020



Abstract:Echocardiogram (echo) is the earliest and the primary tool for identifying regional wall motion abnormalities (RWMA) in order to diagnose myocardial infarction (MI) or commonly known as heart attack. This paper proposes a novel approach, Active Polynomials, which can accurately and robustly estimate the global motion of the Left Ventricular (LV) wall from any echo in a robust and accurate way. The proposed algorithm quantifies the true wall motion occurring in LV wall segments so as to assist cardiologists diagnose early signs of an acute MI. It further enables medical experts to gain an enhanced visualization capability of echo images through color-coded segments along with their "maximum motion displacement" plots helping them to better assess wall motion and LV Ejection-Fraction (LVEF). The outputs of the method can further help echo-technicians to assess and improve the quality of the echocardiogram recording. A major contribution of this study is the first public echo database collection composed by physicians at the Hamad Medical Corporation Hospital in Qatar. The so-called HMC-QU database will serve as the benchmark for the forthcoming relevant studies. The results over the HMC-QU dataset show that the proposed approach can achieve high accuracy, sensitivity and precision in MI detection even though the echo quality is quite poor, and the temporal resolution is low.
Reliable Tuberculosis Detection using Chest X-ray with Deep Learning, Segmentation and Visualization
Jul 29, 2020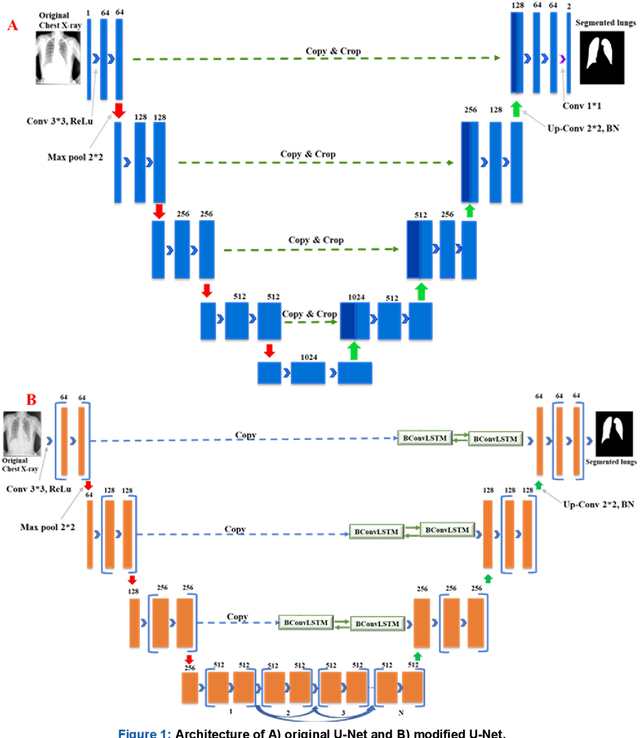
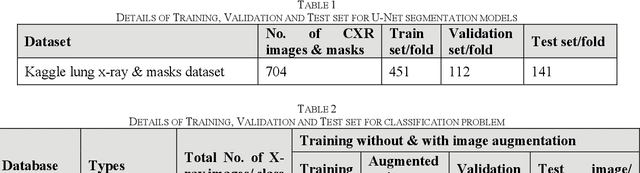
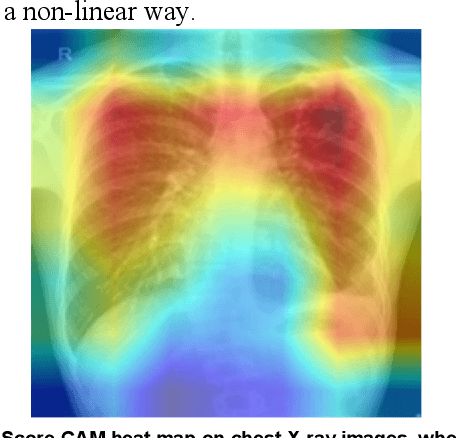
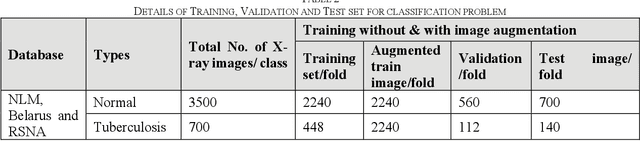
Abstract:Tuberculosis (TB) is a chronic lung disease that occurs due to bacterial infection and is one of the top 10 leading causes of death. Accurate and early detection of TB is very important, otherwise, it could be life-threatening. In this work, we have detected TB reliably from the chest X-ray images using image pre-processing, data augmentation, image segmentation, and deep-learning classification techniques. Several public databases were used to create a database of 700 TB infected and 3500 normal chest X-ray images for this study. Nine different deep CNNs (ResNet18, ResNet50, ResNet101, ChexNet, InceptionV3, Vgg19, DenseNet201, SqueezeNet, and MobileNet), which were used for transfer learning from their pre-trained initial weights and trained, validated and tested for classifying TB and non-TB normal cases. Three different experiments were carried out in this work: segmentation of X-ray images using two different U-net models, classification using X-ray images, and segmented lung images. The accuracy, precision, sensitivity, F1-score, specificity in the detection of tuberculosis using X-ray images were 97.07 %, 97.34 %, 97.07 %, 97.14 % and 97.36 % respectively. However, segmented lungs for the classification outperformed than whole X-ray image-based classification and accuracy, precision, sensitivity, F1-score, specificity were 99.9 %, 99.91 %, 99.9 %, 99.9 %, and 99.52 % respectively. The paper also used a visualization technique to confirm that CNN learns dominantly from the segmented lung regions results in higher detection accuracy. The proposed method with state-of-the-art performance can be useful in the computer-aided faster diagnosis of tuberculosis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge