Rushi Lan
FakeRadar: Probing Forgery Outliers to Detect Unknown Deepfake Videos
Dec 16, 2025Abstract:In this paper, we propose FakeRadar, a novel deepfake video detection framework designed to address the challenges of cross-domain generalization in real-world scenarios. Existing detection methods typically rely on manipulation-specific cues, performing well on known forgery types but exhibiting severe limitations against emerging manipulation techniques. This poor generalization stems from their inability to adapt effectively to unseen forgery patterns. To overcome this, we leverage large-scale pretrained models (e.g. CLIP) to proactively probe the feature space, explicitly highlighting distributional gaps between real videos, known forgeries, and unseen manipulations. Specifically, FakeRadar introduces Forgery Outlier Probing, which employs dynamic subcluster modeling and cluster-conditional outlier generation to synthesize outlier samples near boundaries of estimated subclusters, simulating novel forgery artifacts beyond known manipulation types. Additionally, we design Outlier-Guided Tri-Training, which optimizes the detector to distinguish real, fake, and outlier samples using proposed outlier-driven contrastive learning and outlier-conditioned cross-entropy losses. Experiments show that FakeRadar outperforms existing methods across various benchmark datasets for deepfake video detection, particularly in cross-domain evaluations, by handling the variety of emerging manipulation techniques.
Lightweight Joint Audio-Visual Deepfake Detection via Single-Stream Multi-Modal Learning Framework
Jun 09, 2025Abstract:Deepfakes are AI-synthesized multimedia data that may be abused for spreading misinformation. Deepfake generation involves both visual and audio manipulation. To detect audio-visual deepfakes, previous studies commonly employ two relatively independent sub-models to learn audio and visual features, respectively, and fuse them subsequently for deepfake detection. However, this may underutilize the inherent correlations between audio and visual features. Moreover, utilizing two isolated feature learning sub-models can result in redundant neural layers, making the overall model inefficient and impractical for resource-constrained environments. In this work, we design a lightweight network for audio-visual deepfake detection via a single-stream multi-modal learning framework. Specifically, we introduce a collaborative audio-visual learning block to efficiently integrate multi-modal information while learning the visual and audio features. By iteratively employing this block, our single-stream network achieves a continuous fusion of multi-modal features across its layers. Thus, our network efficiently captures visual and audio features without the need for excessive block stacking, resulting in a lightweight network design. Furthermore, we propose a multi-modal classification module that can boost the dependence of the visual and audio classifiers on modality content. It also enhances the whole resistance of the video classifier against the mismatches between audio and visual modalities. We conduct experiments on the DF-TIMIT, FakeAVCeleb, and DFDC benchmark datasets. Compared to state-of-the-art audio-visual joint detection methods, our method is significantly lightweight with only 0.48M parameters, yet it achieves superiority in both uni-modal and multi-modal deepfakes, as well as in unseen types of deepfakes.
Phoneme-Level Feature Discrepancies: A Key to Detecting Sophisticated Speech Deepfakes
Dec 17, 2024



Abstract:Recent advancements in text-to-speech and speech conversion technologies have enabled the creation of highly convincing synthetic speech. While these innovations offer numerous practical benefits, they also cause significant security challenges when maliciously misused. Therefore, there is an urgent need to detect these synthetic speech signals. Phoneme features provide a powerful speech representation for deepfake detection. However, previous phoneme-based detection approaches typically focused on specific phonemes, overlooking temporal inconsistencies across the entire phoneme sequence. In this paper, we develop a new mechanism for detecting speech deepfakes by identifying the inconsistencies of phoneme-level speech features. We design an adaptive phoneme pooling technique that extracts sample-specific phoneme-level features from frame-level speech data. By applying this technique to features extracted by pre-trained audio models on previously unseen deepfake datasets, we demonstrate that deepfake samples often exhibit phoneme-level inconsistencies when compared to genuine speech. To further enhance detection accuracy, we propose a deepfake detector that uses a graph attention network to model the temporal dependencies of phoneme-level features. Additionally, we introduce a random phoneme substitution augmentation technique to increase feature diversity during training. Extensive experiments on four benchmark datasets demonstrate the superior performance of our method over existing state-of-the-art detection methods.
Gland segmentation via dual encoders and boundary-enhanced attention
Jan 29, 2024



Abstract:Accurate and automated gland segmentation on pathological images can assist pathologists in diagnosing the malignancy of colorectal adenocarcinoma. However, due to various gland shapes, severe deformation of malignant glands, and overlapping adhesions between glands. Gland segmentation has always been very challenging. To address these problems, we propose a DEA model. This model consists of two branches: the backbone encoding and decoding network and the local semantic extraction network. The backbone encoding and decoding network extracts advanced Semantic features, uses the proposed feature decoder to restore feature space information, and then enhances the boundary features of the gland through boundary enhancement attention. The local semantic extraction network uses the pre-trained DeepLabv3+ as a Local semantic-guided encoder to realize the extraction of edge features. Experimental results on two public datasets, GlaS and CRAG, confirm that the performance of our method is better than other gland segmentation methods.
A novel dataset and a two-stage mitosis nuclei detection method based on hybrid anchor branch
Jan 18, 2023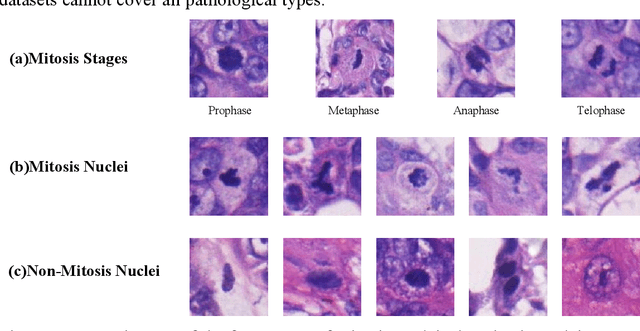
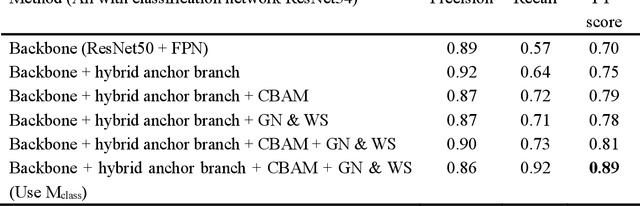
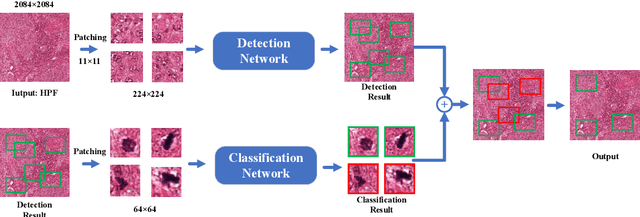
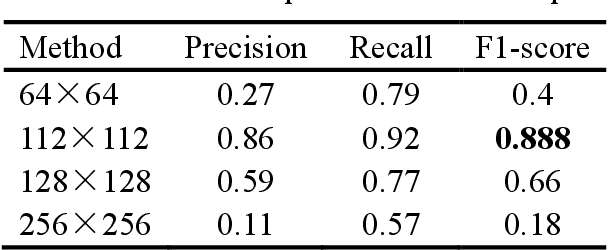
Abstract:Mitosis detection is one of the challenging problems in computational pathology, and mitotic count is an important index of cancer grading for pathologists. However, current counts of mitotic nuclei rely on pathologists looking microscopically at the number of mitotic nuclei in hot spots, which is subjective and time-consuming. In this paper, we propose a two-stage cascaded network, named FoCasNet, for mitosis detection. In the first stage, a detection network named M_det is proposed to detect as many mitoses as possible. In the second stage, a classification network M_class is proposed to refine the results of the first stage. In addition, the attention mechanism, normalization method, and hybrid anchor branch classification subnet are introduced to improve the overall detection performance. Our method achieves the current highest F1-score of 0.888 on the public dataset ICPR 2012. We also evaluated our method on the GZMH dataset released by our research team for the first time and reached the highest F1-score of 0.563, which is also better than multiple classic detection networks widely used at present. It confirmed the effectiveness and generalization of our method. The code will be available at: https://github.com/antifen/mitosis-nuclei-detection.
A Novel Dataset and a Deep Learning Method for Mitosis Nuclei Segmentation and Classification
Dec 27, 2022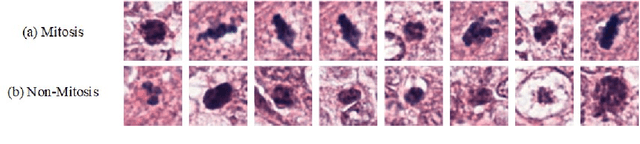
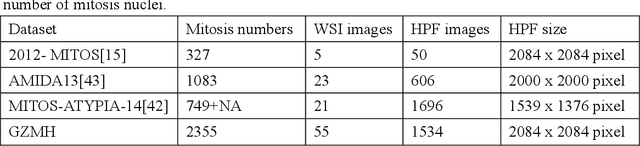
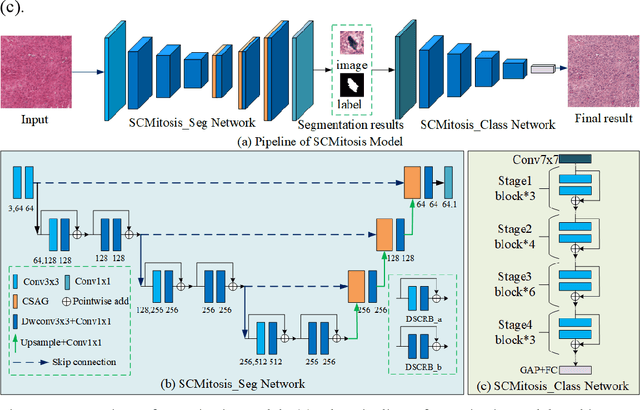
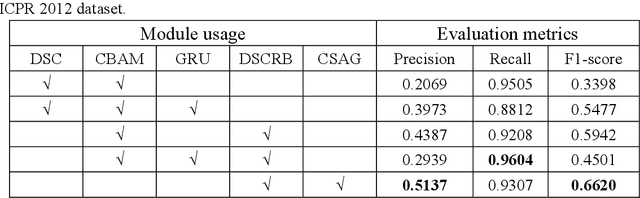
Abstract:Mitosis nuclei count is one of the important indicators for the pathological diagnosis of breast cancer. The manual annotation needs experienced pathologists, which is very time-consuming and inefficient. With the development of deep learning methods, some models with good performance have emerged, but the generalization ability should be further strengthened. In this paper, we propose a two-stage mitosis segmentation and classification method, named SCMitosis. Firstly, the segmentation performance with a high recall rate is achieved by the proposed depthwise separable convolution residual block and channel-spatial attention gate. Then, a classification network is cascaded to further improve the detection performance of mitosis nuclei. The proposed model is verified on the ICPR 2012 dataset, and the highest F-score value of 0.8687 is obtained compared with the current state-of-the-art algorithms. In addition, the model also achieves good performance on GZMH dataset, which is prepared by our group and will be firstly released with the publication of this paper. The code will be available at: https://github.com/antifen/mitosis-nuclei-segmentation.
Binary Representation via Jointly Personalized Sparse Hashing
Aug 31, 2022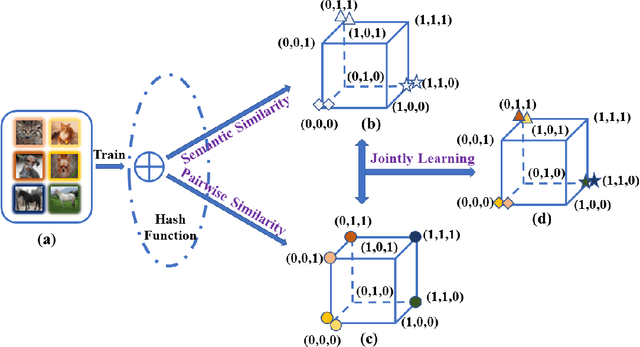
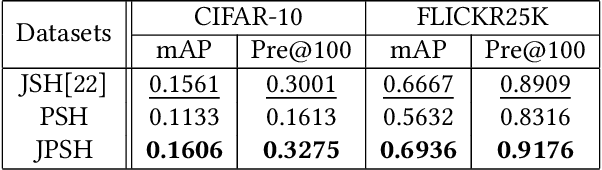
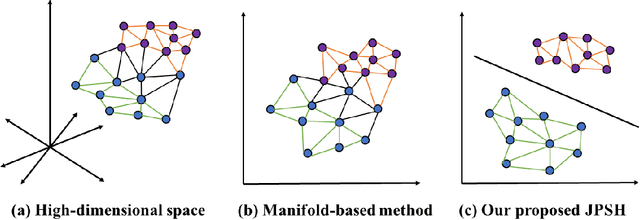
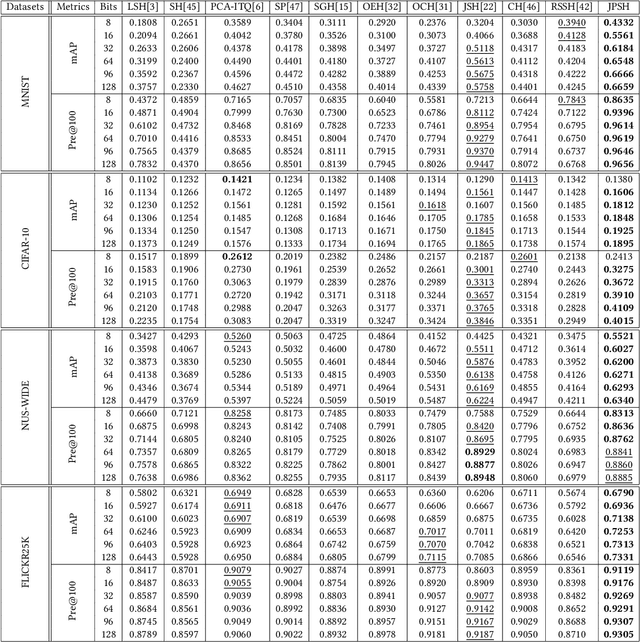
Abstract:Unsupervised hashing has attracted much attention for binary representation learning due to the requirement of economical storage and efficiency of binary codes. It aims to encode high-dimensional features in the Hamming space with similarity preservation between instances. However, most existing methods learn hash functions in manifold-based approaches. Those methods capture the local geometric structures (i.e., pairwise relationships) of data, and lack satisfactory performance in dealing with real-world scenarios that produce similar features (e.g. color and shape) with different semantic information. To address this challenge, in this work, we propose an effective unsupervised method, namely Jointly Personalized Sparse Hashing (JPSH), for binary representation learning. To be specific, firstly, we propose a novel personalized hashing module, i.e., Personalized Sparse Hashing (PSH). Different personalized subspaces are constructed to reflect category-specific attributes for different clusters, adaptively mapping instances within the same cluster to the same Hamming space. In addition, we deploy sparse constraints for different personalized subspaces to select important features. We also collect the strengths of the other clusters to build the PSH module with avoiding over-fitting. Then, to simultaneously preserve semantic and pairwise similarities in our JPSH, we incorporate the PSH and manifold-based hash learning into the seamless formulation. As such, JPSH not only distinguishes the instances from different clusters, but also preserves local neighborhood structures within the cluster. Finally, an alternating optimization algorithm is adopted to iteratively capture analytical solutions of the JPSH model. Extensive experiments on four benchmark datasets verify that the JPSH outperforms several hashing algorithms on the similarity search task.
AIM 2020 Challenge on Efficient Super-Resolution: Methods and Results
Sep 15, 2020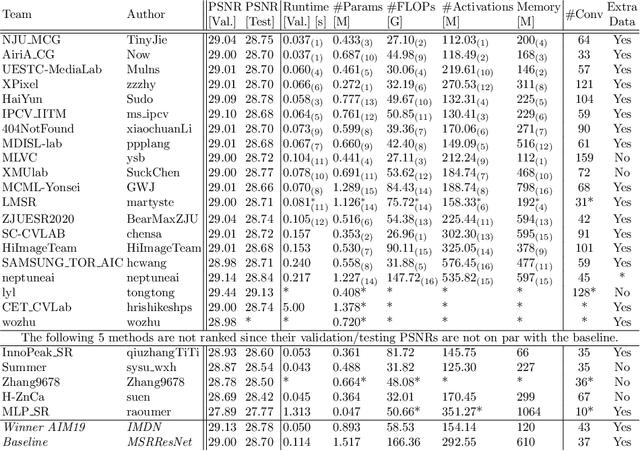
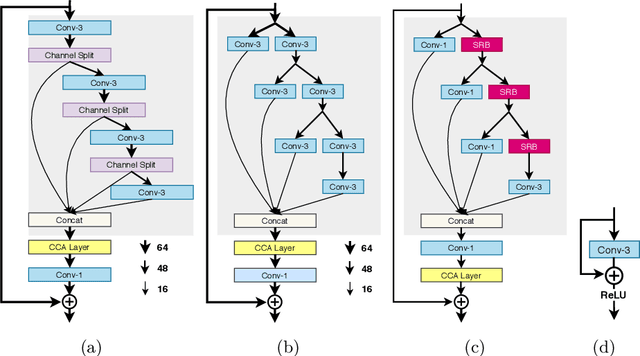
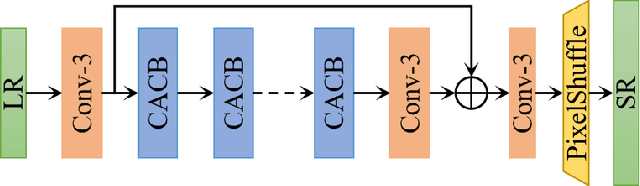
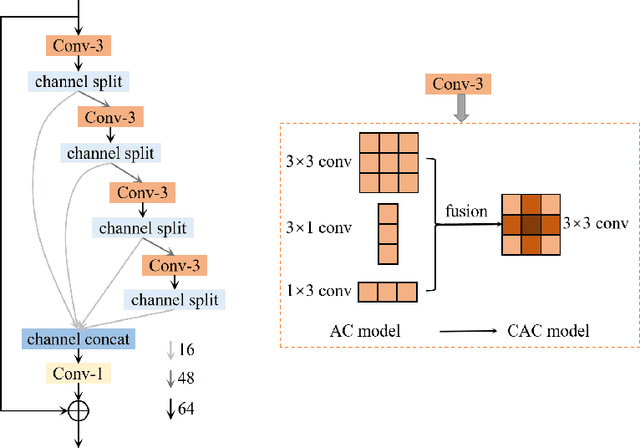
Abstract:This paper reviews the AIM 2020 challenge on efficient single image super-resolution with focus on the proposed solutions and results. The challenge task was to super-resolve an input image with a magnification factor x4 based on a set of prior examples of low and corresponding high resolution images. The goal is to devise a network that reduces one or several aspects such as runtime, parameter count, FLOPs, activations, and memory consumption while at least maintaining PSNR of MSRResNet. The track had 150 registered participants, and 25 teams submitted the final results. They gauge the state-of-the-art in efficient single image super-resolution.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge