Zhipeng Liu
VC-Bench: Pioneering the Video Connecting Benchmark with a Dataset and Evaluation Metrics
Jan 27, 2026Abstract:While current video generation focuses on text or image conditions, practical applications like video editing and vlogging often need to seamlessly connect separate clips. In our work, we introduce Video Connecting, an innovative task that aims to generate smooth intermediate video content between given start and end clips. However, the absence of standardized evaluation benchmarks has hindered the development of this task. To bridge this gap, we proposed VC-Bench, a novel benchmark specifically designed for video connecting. It includes 1,579 high-quality videos collected from public platforms, covering 15 main categories and 72 subcategories to ensure diversity and structure. VC-Bench focuses on three core aspects: Video Quality Score VQS, Start-End Consistency Score SECS, and Transition Smoothness Score TSS. Together, they form a comprehensive framework that moves beyond conventional quality-only metrics. We evaluated multiple state-of-the-art video generation models on VC-Bench. Experimental results reveal significant limitations in maintaining start-end consistency and transition smoothness, leading to lower overall coherence and fluidity. We expect that VC-Bench will serve as a pioneering benchmark to inspire and guide future research in video connecting. The evaluation metrics and dataset are publicly available at: https://anonymous.4open.science/r/VC-Bench-1B67/.
Beyond Visual Realism: Toward Reliable Financial Time Series Generation
Jan 19, 2026Abstract:Generative models for financial time series often create data that look realistic and even reproduce stylized facts such as fat tails or volatility clustering. However, these apparent successes break down under trading backtests: models like GANs or WGAN-GP frequently collapse, yielding extreme and unrealistic results that make the synthetic data unusable in practice. We identify the root cause in the neglect of financial asymmetry and rare tail events, which strongly affect market risk but are often overlooked by objectives focusing on distribution matching. To address this, we introduce the Stylized Facts Alignment GAN (SFAG), which converts key stylized facts into differentiable structural constraints and jointly optimizes them with adversarial loss. This multi-constraint design ensures that generated series remain aligned with market dynamics not only in plots but also in backtesting. Experiments on the Shanghai Composite Index (2004--2024) show that while baseline GANs produce unstable and implausible trading outcomes, SFAG generates synthetic data that preserve stylized facts and support robust momentum strategy performance. Our results highlight that structure-preserving objectives are essential to bridge the gap between superficial realism and practical usability in financial generative modeling.
We Need a More Robust Classifier: Dual Causal Learning Empowers Domain-Incremental Time Series Classification
Jan 15, 2026Abstract:The World Wide Web thrives on intelligent services that rely on accurate time series classification, which has recently witnessed significant progress driven by advances in deep learning. However, existing studies face challenges in domain incremental learning. In this paper, we propose a lightweight and robust dual-causal disentanglement framework (DualCD) to enhance the robustness of models under domain incremental scenarios, which can be seamlessly integrated into time series classification models. Specifically, DualCD first introduces a temporal feature disentanglement module to capture class-causal features and spurious features. The causal features can offer sufficient predictive power to support the classifier in domain incremental learning settings. To accurately capture these causal features, we further design a dual-causal intervention mechanism to eliminate the influence of both intra-class and inter-class confounding features. This mechanism constructs variant samples by combining the current class's causal features with intra-class spurious features and with causal features from other classes. The causal intervention loss encourages the model to accurately predict the labels of these variant samples based solely on the causal features. Extensive experiments on multiple datasets and models demonstrate that DualCD effectively improves performance in domain incremental scenarios. We summarize our rich experiments into a comprehensive benchmark to facilitate research in domain incremental time series classification.
CogniSNN: Enabling Neuron-Expandability, Pathway-Reusability, and Dynamic-Configurability with Random Graph Architectures in Spiking Neural Networks
Dec 12, 2025Abstract:Spiking neural networks (SNNs), regarded as the third generation of artificial neural networks, are expected to bridge the gap between artificial intelligence and computational neuroscience. However, most mainstream SNN research directly adopts the rigid, chain-like hierarchical architecture of traditional artificial neural networks (ANNs), ignoring key structural characteristics of the brain. Biological neurons are stochastically interconnected, forming complex neural pathways that exhibit Neuron-Expandability, Pathway-Reusability, and Dynamic-Configurability. In this paper, we introduce a new SNN paradigm, named Cognition-aware SNN (CogniSNN), by incorporating Random Graph Architecture (RGA). Furthermore, we address the issues of network degradation and dimensional mismatch in deep pathways by introducing an improved pure spiking residual mechanism alongside an adaptive pooling strategy. Then, we design a Key Pathway-based Learning without Forgetting (KP-LwF) approach, which selectively reuses critical neural pathways while retaining historical knowledge, enabling efficient multi-task transfer. Finally, we propose a Dynamic Growth Learning (DGL) algorithm that allows neurons and synapses to grow dynamically along the internal temporal dimension. Extensive experiments demonstrate that CogniSNN achieves performance comparable to, or even surpassing, current state-of-the-art SNNs on neuromorphic datasets and Tiny-ImageNet. The Pathway-Reusability enhances the network's continuous learning capability across different scenarios, while the dynamic growth algorithm improves robustness against interference and mitigates the fixed-timestep constraints during neuromorphic chip deployment. This work demonstrates the potential of SNNs with random graph structures in advancing brain-inspired intelligence and lays the foundation for their practical application on neuromorphic hardware.
CogniSNN: A First Exploration to Random Graph Architecture based Spiking Neural Networks with Enhanced Expandability and Neuroplasticity
May 09, 2025



Abstract:Despite advances in spiking neural networks (SNNs) in numerous tasks, their architectures remain highly similar to traditional artificial neural networks (ANNs), restricting their ability to mimic natural connections between biological neurons. This paper develops a new modeling paradigm for SNN with random graph architecture (RGA), termed Cognition-aware SNN (CogniSNN). Furthermore, we improve the expandability and neuroplasticity of CogniSNN by introducing a modified spiking residual neural node (ResNode) to counteract network degradation in deeper graph pathways, as well as a critical path-based algorithm that enables CogniSNN to perform continual learning on new tasks leveraging the features of the data and the RGA learned in the old task. Experiments show that CogniSNN with re-designed ResNode performs outstandingly in neuromorphic datasets with fewer parameters, achieving 95.5% precision in the DVS-Gesture dataset with only 5 timesteps. The critical path-based approach decreases 3% to 5% forgetting while maintaining expected performance in learning new tasks that are similar to or distinct from the old ones. This study showcases the potential of RGA-based SNN and paves a new path for biologically inspired networks based on graph theory.
CISum: Learning Cross-modality Interaction to Enhance Multimodal Semantic Coverage for Multimodal Summarization
Feb 20, 2023Abstract:Multimodal summarization (MS) aims to generate a summary from multimodal input. Previous works mainly focus on textual semantic coverage metrics such as ROUGE, which considers the visual content as supplemental data. Therefore, the summary is ineffective to cover the semantics of different modalities. This paper proposes a multi-task cross-modality learning framework (CISum) to improve multimodal semantic coverage by learning the cross-modality interaction in the multimodal article. To obtain the visual semantics, we translate images into visual descriptions based on the correlation with text content. Then, the visual description and text content are fused to generate the textual summary to capture the semantics of the multimodal content, and the most relevant image is selected as the visual summary. Furthermore, we design an automatic multimodal semantics coverage metric to evaluate the performance. Experimental results show that CISum outperforms baselines in multimodal semantics coverage metrics while maintaining the excellent performance of ROUGE and BLEU.
A Novel Dataset and a Deep Learning Method for Mitosis Nuclei Segmentation and Classification
Dec 27, 2022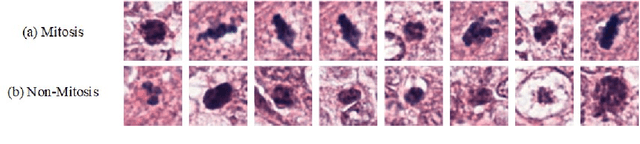
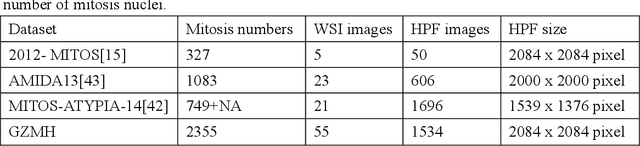
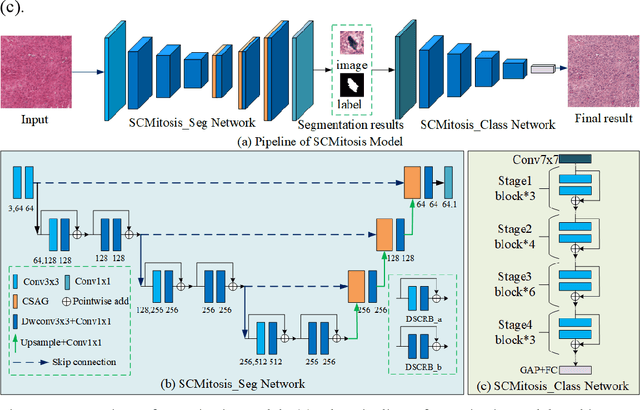
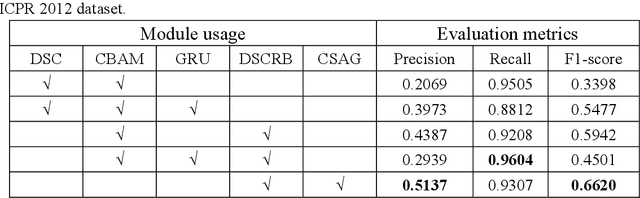
Abstract:Mitosis nuclei count is one of the important indicators for the pathological diagnosis of breast cancer. The manual annotation needs experienced pathologists, which is very time-consuming and inefficient. With the development of deep learning methods, some models with good performance have emerged, but the generalization ability should be further strengthened. In this paper, we propose a two-stage mitosis segmentation and classification method, named SCMitosis. Firstly, the segmentation performance with a high recall rate is achieved by the proposed depthwise separable convolution residual block and channel-spatial attention gate. Then, a classification network is cascaded to further improve the detection performance of mitosis nuclei. The proposed model is verified on the ICPR 2012 dataset, and the highest F-score value of 0.8687 is obtained compared with the current state-of-the-art algorithms. In addition, the model also achieves good performance on GZMH dataset, which is prepared by our group and will be firstly released with the publication of this paper. The code will be available at: https://github.com/antifen/mitosis-nuclei-segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge