Nikolaos Evangelou
A Mechanistic Analysis of Transformers for Dynamical Systems
Dec 24, 2025Abstract:Transformers are increasingly adopted for modeling and forecasting time-series, yet their internal mechanisms remain poorly understood from a dynamical systems perspective. In contrast to classical autoregressive and state-space models, which benefit from well-established theoretical foundations, Transformer architectures are typically treated as black boxes. This gap becomes particularly relevant as attention-based models are considered for general-purpose or zero-shot forecasting across diverse dynamical regimes. In this work, we do not propose a new forecasting model, but instead investigate the representational capabilities and limitations of single-layer Transformers when applied to dynamical data. Building on a dynamical systems perspective we interpret causal self-attention as a linear, history-dependent recurrence and analyze how it processes temporal information. Through a series of linear and nonlinear case studies, we identify distinct operational regimes. For linear systems, we show that the convexity constraint imposed by softmax attention fundamentally restricts the class of dynamics that can be represented, leading to oversmoothing in oscillatory settings. For nonlinear systems under partial observability, attention instead acts as an adaptive delay-embedding mechanism, enabling effective state reconstruction when sufficient temporal context and latent dimensionality are available. These results help bridge empirical observations with classical dynamical systems theory, providing insight into when and why Transformers succeed or fail as models of dynamical systems.
Generative Learning for Slow Manifolds and Bifurcation Diagrams
Apr 29, 2025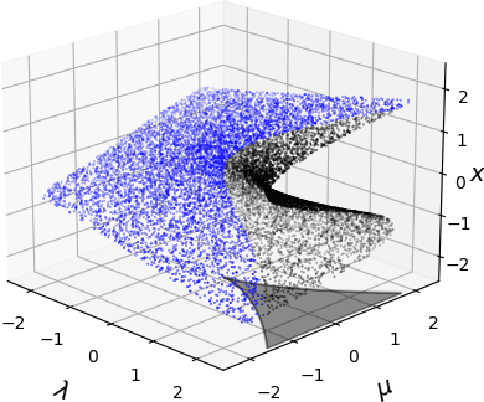
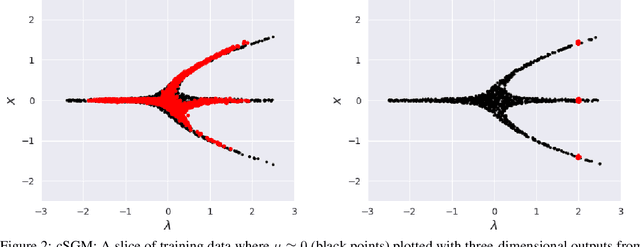
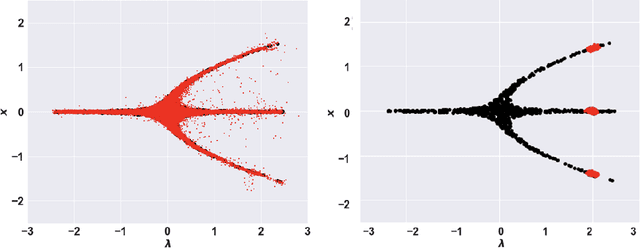
Abstract:In dynamical systems characterized by separation of time scales, the approximation of so called ``slow manifolds'', on which the long term dynamics lie, is a useful step for model reduction. Initializing on such slow manifolds is a useful step in modeling, since it circumvents fast transients, and is crucial in multiscale algorithms alternating between fine scale (fast) and coarser scale (slow) simulations. In a similar spirit, when one studies the infinite time dynamics of systems depending on parameters, the system attractors (e.g., its steady states) lie on bifurcation diagrams. Sampling these manifolds gives us representative attractors (here, steady states of ODEs or PDEs) at different parameter values. Algorithms for the systematic construction of these manifolds are required parts of the ``traditional'' numerical nonlinear dynamics toolkit. In more recent years, as the field of Machine Learning develops, conditional score-based generative models (cSGMs) have demonstrated capabilities in generating plausible data from target distributions that are conditioned on some given label. It is tempting to exploit such generative models to produce samples of data distributions conditioned on some quantity of interest (QoI). In this work, we present a framework for using cSGMs to quickly (a) initialize on a low-dimensional (reduced-order) slow manifold of a multi-time-scale system consistent with desired value(s) of a QoI (a ``label'') on the manifold, and (b) approximate steady states in a bifurcation diagram consistent with a (new, out-of-sample) parameter value. This conditional sampling can help uncover the geometry of the reduced slow-manifold and/or approximately ``fill in'' missing segments of steady states in a bifurcation diagram.
Comparing analytic and data-driven approaches to parameter identifiability: A power systems case study
Dec 24, 2024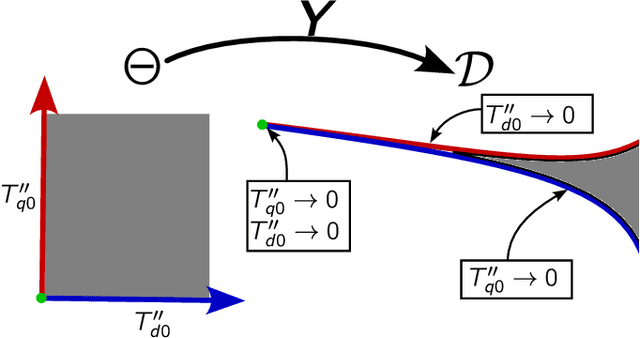
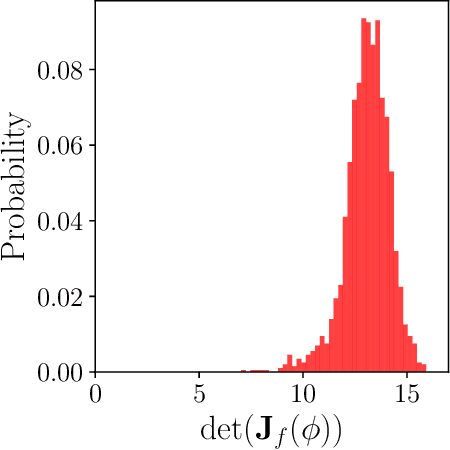
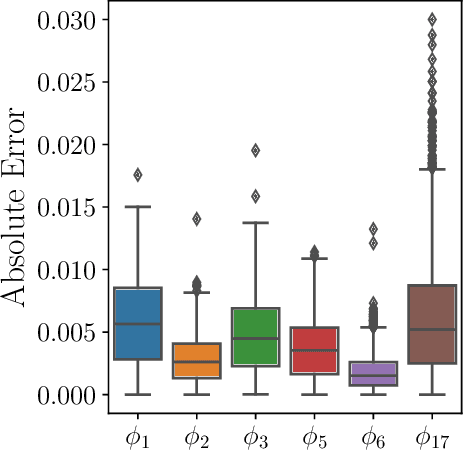
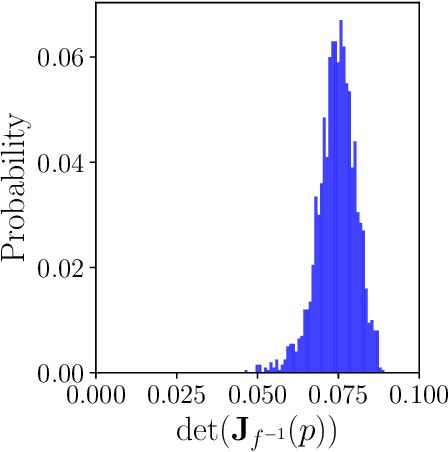
Abstract:Parameter identifiability refers to the capability of accurately inferring the parameter values of a model from its observations (data). Traditional analysis methods exploit analytical properties of the closed form model, in particular sensitivity analysis, to quantify the response of the model predictions to variations in parameters. Techniques developed to analyze data, specifically manifold learning methods, have the potential to complement, and even extend the scope of the traditional analytical approaches. We report on a study comparing and contrasting analytical and data-driven approaches to quantify parameter identifiability and, importantly, perform parameter reduction tasks. We use the infinite bus synchronous generator model, a well-understood model from the power systems domain, as our benchmark problem. Our traditional analysis methods use the Fisher Information Matrix to quantify parameter identifiability analysis, and the Manifold Boundary Approximation Method to perform parameter reduction. We compare these results to those arrived at through data-driven manifold learning schemes: Output - Diffusion Maps and Geometric Harmonics. For our test case, we find that the two suites of tools (analytical when a model is explicitly available, as well as data-driven when the model is lacking and only measurement data are available) give (correct) comparable results; these results are also in agreement with traditional analysis based on singular perturbation theory. We then discuss the prospects of using data-driven methods for such model analysis.
From Noise to Signal: Unveiling Treatment Effects from Digital Health Data through Pharmacology-Informed Neural-SDE
Mar 05, 2024



Abstract:Digital health technologies (DHT), such as wearable devices, provide personalized, continuous, and real-time monitoring of patient. These technologies are contributing to the development of novel therapies and personalized medicine. Gaining insight from these technologies requires appropriate modeling techniques to capture clinically-relevant changes in disease state. The data generated from these devices is characterized by being stochastic in nature, may have missing elements, and exhibits considerable inter-individual variability - thereby making it difficult to analyze using traditional longitudinal modeling techniques. We present a novel pharmacology-informed neural stochastic differential equation (SDE) model capable of addressing these challenges. Using synthetic data, we demonstrate that our approach is effective in identifying treatment effects and learning causal relationships from stochastic data, thereby enabling counterfactual simulation.
Tipping Points of Evolving Epidemiological Networks: Machine Learning-Assisted, Data-Driven Effective Modeling
Nov 10, 2023



Abstract:We study the tipping point collective dynamics of an adaptive susceptible-infected-susceptible (SIS) epidemiological network in a data-driven, machine learning-assisted manner. We identify a parameter-dependent effective stochastic differential equation (eSDE) in terms of physically meaningful coarse mean-field variables through a deep-learning ResNet architecture inspired by numerical stochastic integrators. We construct an approximate effective bifurcation diagram based on the identified drift term of the eSDE and contrast it with the mean-field SIS model bifurcation diagram. We observe a subcritical Hopf bifurcation in the evolving network's effective SIS dynamics, that causes the tipping point behavior; this takes the form of large amplitude collective oscillations that spontaneously -- yet rarely -- arise from the neighborhood of a (noisy) stationary state. We study the statistics of these rare events both through repeated brute force simulations and by using established mathematical/computational tools exploiting the right-hand-side of the identified SDE. We demonstrate that such a collective SDE can also be identified (and the rare events computations also performed) in terms of data-driven coarse observables, obtained here via manifold learning techniques, in particular Diffusion Maps. The workflow of our study is straightforwardly applicable to other complex dynamics problems exhibiting tipping point dynamics.
Machine Learning for the identification of phase-transitions in interacting agent-based systems
Oct 29, 2023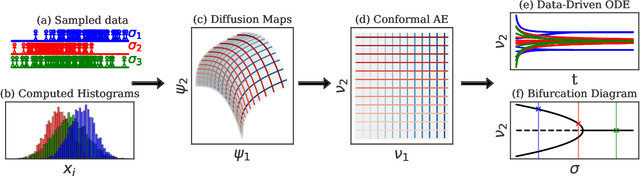
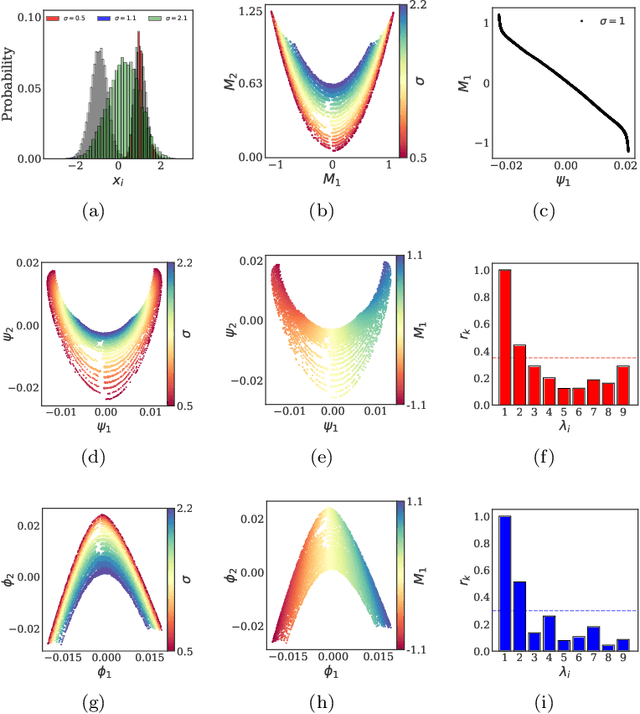
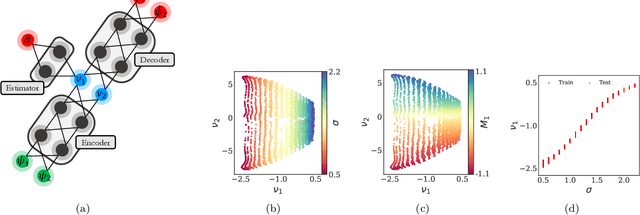
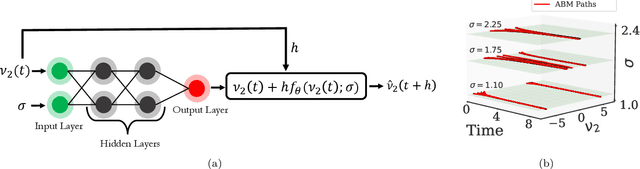
Abstract:Deriving closed-form, analytical expressions for reduced-order models, and judiciously choosing the closures leading to them, has long been the strategy of choice for studying phase- and noise-induced transitions for agent-based models (ABMs). In this paper, we propose a data-driven framework that pinpoints phase transitions for an ABM in its mean-field limit, using a smaller number of variables than traditional closed-form models. To this end, we use the manifold learning algorithm Diffusion Maps to identify a parsimonious set of data-driven latent variables, and show that they are in one-to-one correspondence with the expected theoretical order parameter of the ABM. We then utilize a deep learning framework to obtain a conformal reparametrization of the data-driven coordinates that facilitates, in our example, the identification of a single parameter-dependent ODE in these coordinates. We identify this ODE through a residual neural network inspired by a numerical integration scheme (forward Euler). We then use the identified ODE -- enabled through an odd symmetry transformation -- to construct the bifurcation diagram exhibiting the phase transition.
Nonlinear dimensionality reduction then and now: AIMs for dissipative PDEs in the ML era
Oct 24, 2023



Abstract:This study presents a collection of purely data-driven workflows for constructing reduced-order models (ROMs) for distributed dynamical systems. The ROMs we focus on, are data-assisted models inspired by, and templated upon, the theory of Approximate Inertial Manifolds (AIMs); the particular motivation is the so-called post-processing Galerkin method of Garcia-Archilla, Novo and Titi. Its applicability can be extended: the need for accurate truncated Galerkin projections and for deriving closed-formed corrections can be circumvented using machine learning tools. When the right latent variables are not a priori known, we illustrate how autoencoders as well as Diffusion Maps (a manifold learning scheme) can be used to discover good sets of latent variables and test their explainability. The proposed methodology can express the ROMs in terms of (a) theoretical (Fourier coefficients), (b) linear data-driven (POD modes) and/or (c) nonlinear data-driven (Diffusion Maps) coordinates. Both Black-Box and (theoretically-informed and data-corrected) Gray-Box models are described; the necessity for the latter arises when truncated Galerkin projections are so inaccurate as to not be amenable to post-processing. We use the Chafee-Infante reaction-diffusion and the Kuramoto-Sivashinsky dissipative partial differential equations to illustrate and successfully test the overall framework.
Tasks Makyth Models: Machine Learning Assisted Surrogates for Tipping Points
Sep 25, 2023



Abstract:We present a machine learning (ML)-assisted framework bridging manifold learning, neural networks, Gaussian processes, and Equation-Free multiscale modeling, for (a) detecting tipping points in the emergent behavior of complex systems, and (b) characterizing probabilities of rare events (here, catastrophic shifts) near them. Our illustrative example is an event-driven, stochastic agent-based model (ABM) describing the mimetic behavior of traders in a simple financial market. Given high-dimensional spatiotemporal data -- generated by the stochastic ABM -- we construct reduced-order models for the emergent dynamics at different scales: (a) mesoscopic Integro-Partial Differential Equations (IPDEs); and (b) mean-field-type Stochastic Differential Equations (SDEs) embedded in a low-dimensional latent space, targeted to the neighborhood of the tipping point. We contrast the uses of the different models and the effort involved in learning them.
Learning Effective SDEs from Brownian Dynamics Simulations of Colloidal Particles
Apr 30, 2022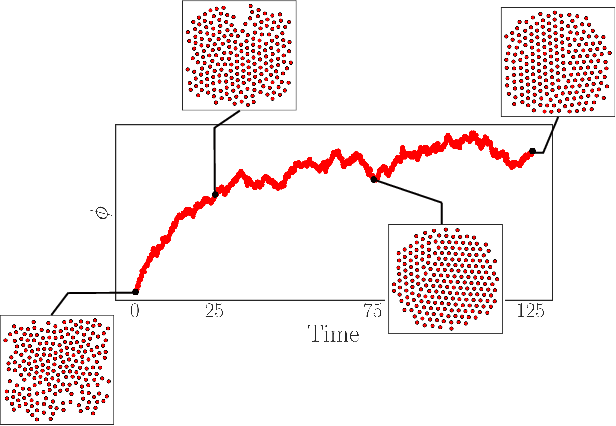
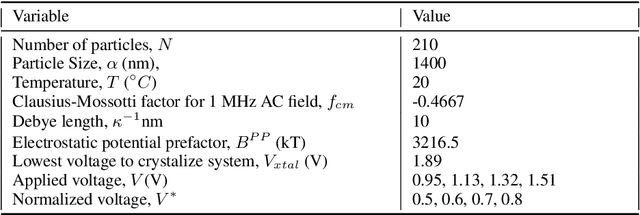
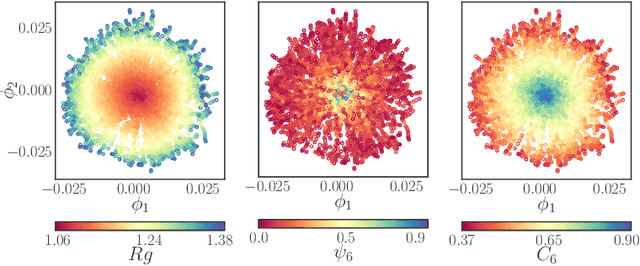
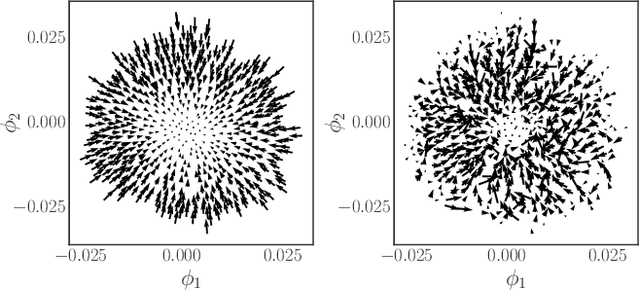
Abstract:We construct a reduced, data-driven, parameter dependent effective Stochastic Differential Equation (eSDE) for electric-field mediated colloidal crystallization using data obtained from Brownian Dynamics Simulations. We use Diffusion Maps (a manifold learning algorithm) to identify a set of useful latent observables. In this latent space we identify an eSDE using a deep learning architecture inspired by numerical stochastic integrators and compare it with the traditional Kramers-Moyal expansion estimation. We show that the obtained variables and the learned dynamics accurately encode the physics of the Brownian Dynamic Simulations. We further illustrate that our reduced model captures the dynamics of corresponding experimental data. Our dimension reduction/reduced model identification approach can be easily ported to a broad class of particle systems dynamics experiments/models.
Double Diffusion Maps and their Latent Harmonics for Scientific Computations in Latent Space
Apr 26, 2022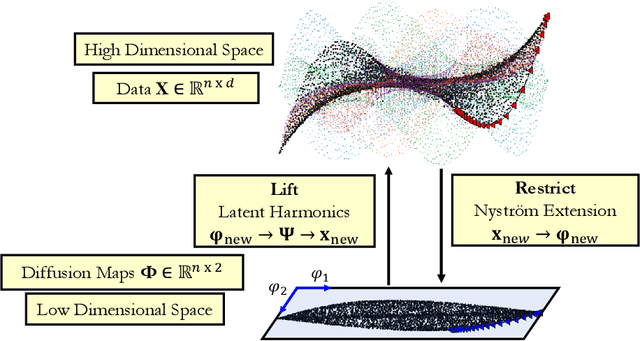


Abstract:We introduce a data-driven approach to building reduced dynamical models through manifold learning; the reduced latent space is discovered using Diffusion Maps (a manifold learning technique) on time series data. A second round of Diffusion Maps on those latent coordinates allows the approximation of the reduced dynamical models. This second round enables mapping the latent space coordinates back to the full ambient space (what is called lifting); it also enables the approximation of full state functions of interest in terms of the reduced coordinates. In our work, we develop and test three different reduced numerical simulation methodologies, either through pre-tabulation in the latent space and integration on the fly or by going back and forth between the ambient space and the latent space. The data-driven latent space simulation results, based on the three different approaches, are validated through (a) the latent space observation of the full simulation through the Nystr\"om Extension formula, or through (b) lifting the reduced trajectory back to the full ambient space, via Latent Harmonics. Latent space modeling often involves additional regularization to favor certain properties of the space over others, and the mapping back to the ambient space is then constructed mostly independently from these properties; here, we use the same data-driven approach to construct the latent space and then map back to the ambient space.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge