Ellis R. Crabtree
Generative Learning for Slow Manifolds and Bifurcation Diagrams
Apr 29, 2025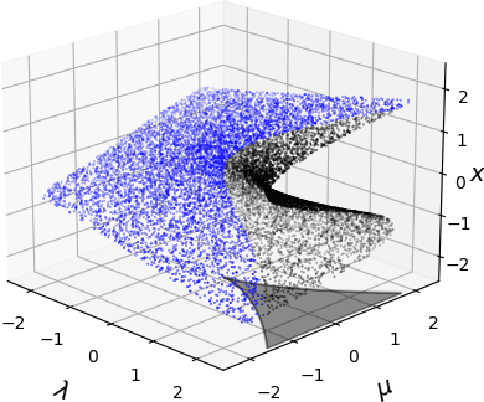
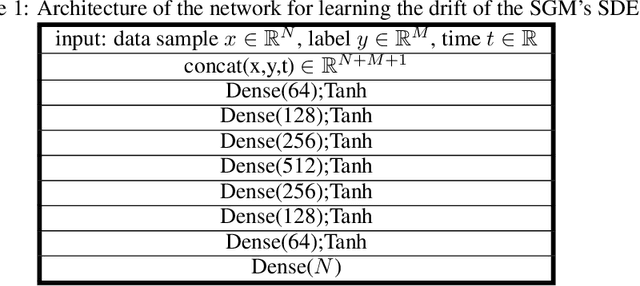
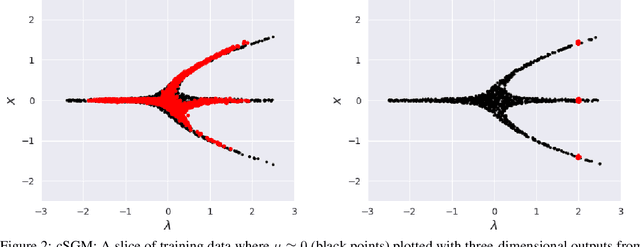
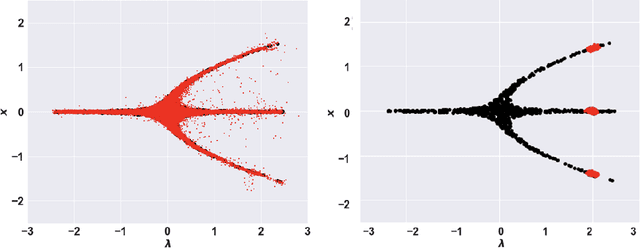
Abstract:In dynamical systems characterized by separation of time scales, the approximation of so called ``slow manifolds'', on which the long term dynamics lie, is a useful step for model reduction. Initializing on such slow manifolds is a useful step in modeling, since it circumvents fast transients, and is crucial in multiscale algorithms alternating between fine scale (fast) and coarser scale (slow) simulations. In a similar spirit, when one studies the infinite time dynamics of systems depending on parameters, the system attractors (e.g., its steady states) lie on bifurcation diagrams. Sampling these manifolds gives us representative attractors (here, steady states of ODEs or PDEs) at different parameter values. Algorithms for the systematic construction of these manifolds are required parts of the ``traditional'' numerical nonlinear dynamics toolkit. In more recent years, as the field of Machine Learning develops, conditional score-based generative models (cSGMs) have demonstrated capabilities in generating plausible data from target distributions that are conditioned on some given label. It is tempting to exploit such generative models to produce samples of data distributions conditioned on some quantity of interest (QoI). In this work, we present a framework for using cSGMs to quickly (a) initialize on a low-dimensional (reduced-order) slow manifold of a multi-time-scale system consistent with desired value(s) of a QoI (a ``label'') on the manifold, and (b) approximate steady states in a bifurcation diagram consistent with a (new, out-of-sample) parameter value. This conditional sampling can help uncover the geometry of the reduced slow-manifold and/or approximately ``fill in'' missing segments of steady states in a bifurcation diagram.
Micro-Macro Consistency in Multiscale Modeling: Score-Based Model Assisted Sampling of Fast/Slow Dynamical Systems
Dec 10, 2023Abstract:A valuable step in the modeling of multiscale dynamical systems in fields such as computational chemistry, biology, materials science and more, is the representative sampling of the phase space over long timescales of interest; this task is not, however, without challenges. For example, the long term behavior of a system with many degrees of freedom often cannot be efficiently computationally explored by direct dynamical simulation; such systems can often become trapped in local free energy minima. In the study of physics-based multi-time-scale dynamical systems, techniques have been developed for enhancing sampling in order to accelerate exploration beyond free energy barriers. On the other hand, in the field of Machine Learning, a generic goal of generative models is to sample from a target density, after training on empirical samples from this density. Score based generative models (SGMs) have demonstrated state-of-the-art capabilities in generating plausible data from target training distributions. Conditional implementations of such generative models have been shown to exhibit significant parallels with long-established -- and physics based -- solutions to enhanced sampling. These physics-based methods can then be enhanced through coupling with the ML generative models, complementing the strengths and mitigating the weaknesses of each technique. In this work, we show that that SGMs can be used in such a coupling framework to improve sampling in multiscale dynamical systems.
GANs and Closures: Micro-Macro Consistency in Multiscale Modeling
Aug 23, 2022



Abstract:Sampling the phase space of molecular systems -- and, more generally, of complex systems effectively modeled by stochastic differential equations -- is a crucial modeling step in many fields, from protein folding to materials discovery. These problems are often multiscale in nature: they can be described in terms of low-dimensional effective free energy surfaces parametrized by a small number of "slow" reaction coordinates; the remaining "fast" degrees of freedom populate an equilibrium measure on the reaction coordinate values. Sampling procedures for such problems are used to estimate effective free energy differences as well as ensemble averages with respect to the conditional equilibrium distributions; these latter averages lead to closures for effective reduced dynamic models. Over the years, enhanced sampling techniques coupled with molecular simulation have been developed. An intriguing analogy arises with the field of Machine Learning (ML), where Generative Adversarial Networks can produce high dimensional samples from low dimensional probability distributions. This sample generation returns plausible high dimensional space realizations of a model state, from information about its low-dimensional representation. In this work, we present an approach that couples physics-based simulations and biasing methods for sampling conditional distributions with ML-based conditional generative adversarial networks for the same task. The "coarse descriptors" on which we condition the fine scale realizations can either be known a priori, or learned through nonlinear dimensionality reduction. We suggest that this may bring out the best features of both approaches: we demonstrate that a framework that couples cGANs with physics-based enhanced sampling techniques can improve multiscale SDE dynamical systems sampling, and even shows promise for systems of increasing complexity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge