Luchen Liu
MASS: Multi-Agent Simulation Scaling for Portfolio Construction
May 15, 2025Abstract:LLM-based multi-agent has gained significant attention for their potential in simulation and enhancing performance. However, existing works are limited to pure simulations or are constrained by predefined workflows, restricting their applicability and effectiveness. In this paper, we introduce the Multi-Agent Scaling Simulation (MASS) for portfolio construction. MASS achieves stable and continuous excess returns by progressively increasing the number of agents for large-scale simulations to gain a superior understanding of the market and optimizing agent distribution end-to-end through a reverse optimization process, rather than relying on a fixed workflow. We demonstrate its superiority through performance experiments, ablation studies, backtesting experiments, experiments on updated data and stock pools, scaling experiments, parameter sensitivity experiments, and visualization experiments, conducted in comparison with 6 state-of-the-art baselines on 3 challenging A-share stock pools. We expect the paradigm established by MASS to expand to other tasks with similar characteristics. The implementation of MASS has been open-sourced at https://github.com/gta0804/MASS.
MMEvalPro: Calibrating Multimodal Benchmarks Towards Trustworthy and Efficient Evaluation
Jun 29, 2024



Abstract:Large Multimodal Models (LMMs) exhibit impressive cross-modal understanding and reasoning abilities, often assessed through multiple-choice questions (MCQs) that include an image, a question, and several options. However, many benchmarks used for such evaluations suffer from systematic biases. Remarkably, Large Language Models (LLMs) without any visual perception capabilities achieve non-trivial performance, undermining the credibility of these evaluations. To address this issue while maintaining the efficiency of MCQ evaluations, we propose MMEvalPro, a benchmark designed to avoid Type-I errors through a trilogy evaluation pipeline and more rigorous metrics. For each original question from existing benchmarks, human annotators augment it by creating one perception question and one knowledge anchor question through a meticulous annotation process. MMEvalPro comprises $2,138$ question triplets, totaling $6,414$ distinct questions. Two-thirds of these questions are manually labeled by human experts, while the rest are sourced from existing benchmarks (MMMU, ScienceQA, and MathVista). Compared with the existing benchmarks, our experiments with the latest LLMs and LMMs demonstrate that MMEvalPro is more challenging (the best LMM lags behind human performance by $31.73\%$, compared to an average gap of $8.03\%$ in previous benchmarks) and more trustworthy (the best LLM trails the best LMM by $23.09\%$, whereas the gap for previous benchmarks is just $14.64\%$). Our in-depth analysis explains the reason for the large performance gap and justifies the trustworthiness of evaluation, underscoring its significant potential for advancing future research.
HIP Network: Historical Information Passing Network for Extrapolation Reasoning on Temporal Knowledge Graph
Feb 19, 2024Abstract:In recent years, temporal knowledge graph (TKG) reasoning has received significant attention. Most existing methods assume that all timestamps and corresponding graphs are available during training, which makes it difficult to predict future events. To address this issue, recent works learn to infer future events based on historical information. However, these methods do not comprehensively consider the latent patterns behind temporal changes, to pass historical information selectively, update representations appropriately and predict events accurately. In this paper, we propose the Historical Information Passing (HIP) network to predict future events. HIP network passes information from temporal, structural and repetitive perspectives, which are used to model the temporal evolution of events, the interactions of events at the same time step, and the known events respectively. In particular, our method considers the updating of relation representations and adopts three scoring functions corresponding to the above dimensions. Experimental results on five benchmark datasets show the superiority of HIP network, and the significant improvements on Hits@1 prove that our method can more accurately predict what is going to happen.
* 7 pages, 3 figures
Redundancy-Free Self-Supervised Relational Learning for Graph Clustering
Sep 09, 2023Abstract:Graph clustering, which learns the node representations for effective cluster assignments, is a fundamental yet challenging task in data analysis and has received considerable attention accompanied by graph neural networks in recent years. However, most existing methods overlook the inherent relational information among the non-independent and non-identically distributed nodes in a graph. Due to the lack of exploration of relational attributes, the semantic information of the graph-structured data fails to be fully exploited which leads to poor clustering performance. In this paper, we propose a novel self-supervised deep graph clustering method named Relational Redundancy-Free Graph Clustering (R$^2$FGC) to tackle the problem. It extracts the attribute- and structure-level relational information from both global and local views based on an autoencoder and a graph autoencoder. To obtain effective representations of the semantic information, we preserve the consistent relation among augmented nodes, whereas the redundant relation is further reduced for learning discriminative embeddings. In addition, a simple yet valid strategy is utilized to alleviate the over-smoothing issue. Extensive experiments are performed on widely used benchmark datasets to validate the superiority of our R$^2$FGC over state-of-the-art baselines. Our codes are available at https://github.com/yisiyu95/R2FGC.
Towards Long-Tailed Recognition for Graph Classification via Collaborative Experts
Sep 05, 2023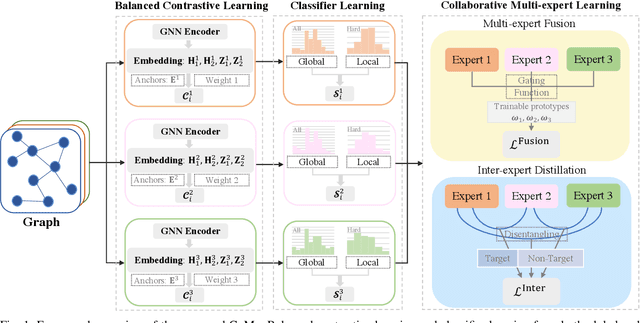
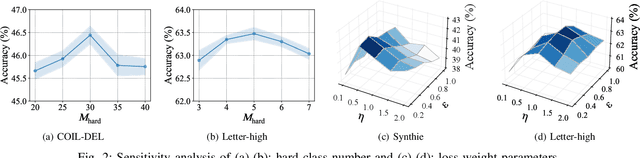
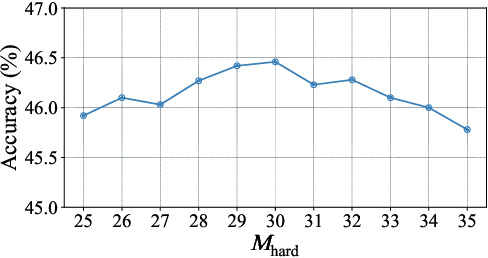
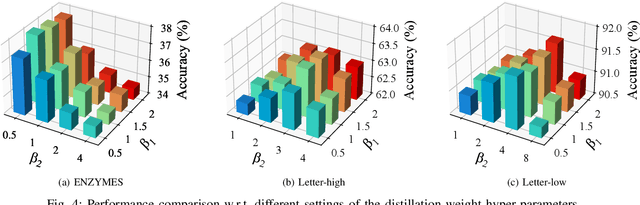
Abstract:Graph classification, aiming at learning the graph-level representations for effective class assignments, has received outstanding achievements, which heavily relies on high-quality datasets that have balanced class distribution. In fact, most real-world graph data naturally presents a long-tailed form, where the head classes occupy much more samples than the tail classes, it thus is essential to study the graph-level classification over long-tailed data while still remaining largely unexplored. However, most existing long-tailed learning methods in visions fail to jointly optimize the representation learning and classifier training, as well as neglect the mining of the hard-to-classify classes. Directly applying existing methods to graphs may lead to sub-optimal performance, since the model trained on graphs would be more sensitive to the long-tailed distribution due to the complex topological characteristics. Hence, in this paper, we propose a novel long-tailed graph-level classification framework via Collaborative Multi-expert Learning (CoMe) to tackle the problem. To equilibrate the contributions of head and tail classes, we first develop balanced contrastive learning from the view of representation learning, and then design an individual-expert classifier training based on hard class mining. In addition, we execute gated fusion and disentangled knowledge distillation among the multiple experts to promote the collaboration in a multi-expert framework. Comprehensive experiments are performed on seven widely-used benchmark datasets to demonstrate the superiority of our method CoMe over state-of-the-art baselines.
Bipartite Graph Embedding via Mutual Information Maximization
Dec 10, 2020



Abstract:Bipartite graph embedding has recently attracted much attention due to the fact that bipartite graphs are widely used in various application domains. Most previous methods, which adopt random walk-based or reconstruction-based objectives, are typically effective to learn local graph structures. However, the global properties of bipartite graph, including community structures of homogeneous nodes and long-range dependencies of heterogeneous nodes, are not well preserved. In this paper, we propose a bipartite graph embedding called BiGI to capture such global properties by introducing a novel local-global infomax objective. Specifically, BiGI first generates a global representation which is composed of two prototype representations. BiGI then encodes sampled edges as local representations via the proposed subgraph-level attention mechanism. Through maximizing the mutual information between local and global representations, BiGI enables nodes in bipartite graph to be globally relevant. Our model is evaluated on various benchmark datasets for the tasks of top-K recommendation and link prediction. Extensive experiments demonstrate that BiGI achieves consistent and significant improvements over state-of-the-art baselines. Detailed analyses verify the high effectiveness of modeling the global properties of bipartite graph.
Multi-task Learning via Adaptation to Similar Tasks for Mortality Prediction of Diverse Rare Diseases
May 11, 2020



Abstract:Mortality prediction of diverse rare diseases using electronic health record (EHR) data is a crucial task for intelligent healthcare. However, data insufficiency and the clinical diversity of rare diseases make it hard for directly training deep learning models on individual disease data or all the data from different diseases. Mortality prediction for these patients with different diseases can be viewed as a multi-task learning problem with insufficient data and large task number. But the tasks with little training data also make it hard to train task-specific modules in multi-task learning models. To address the challenges of data insufficiency and task diversity, we propose an initialization-sharing multi-task learning method (Ada-Sit) which learns the parameter initialization for fast adaptation to dynamically measured similar tasks. We use Ada-Sit to train long short-term memory networks (LSTM) based prediction models on longitudinal EHR data. And experimental results demonstrate that the proposed model is effective for mortality prediction of diverse rare diseases.
Predictive Multi-level Patient Representations from Electronic Health Records
Nov 12, 2019

Abstract:The advent of the Internet era has led to an explosive growth in the Electronic Health Records (EHR) in the past decades. The EHR data can be regarded as a collection of clinical events, including laboratory results, medication records, physiological indicators, etc, which can be used for clinical outcome prediction tasks to support constructions of intelligent health systems. Learning patient representation from these clinical events for the clinical outcome prediction is an important but challenging step. Most related studies transform EHR data of a patient into a sequence of clinical events in temporal order and then use sequential models to learn patient representations for outcome prediction. However, clinical event sequence contains thousands of event types and temporal dependencies. We further make an observation that clinical events occurring in a short period are not constrained by any temporal order but events in a long term are influenced by temporal dependencies. The multi-scale temporal property makes it difficult for traditional sequential models to capture the short-term co-occurrence and the long-term temporal dependencies in clinical event sequences. In response to the above challenges, this paper proposes a Multi-level Representation Model (MRM). MRM first uses a sparse attention mechanism to model the short-term co-occurrence, then uses interval-based event pooling to remove redundant information and reduce sequence length and finally predicts clinical outcomes through Long Short-Term Memory (LSTM). Experiments on real-world datasets indicate that our proposed model largely improves the performance of clinical outcome prediction tasks using EHR data.
Early Prediction of Sepsis From Clinical Datavia Heterogeneous Event Aggregation
Oct 14, 2019



Abstract:Sepsis is a life-threatening condition that seriously endangers millions of people over the world. Hopefully, with the widespread availability of electronic health records (EHR), predictive models that can effectively deal with clinical sequential data increase the possibility to predict sepsis and take early preventive treatment. However, the early prediction is challenging because patients' sequential data in EHR contains temporal interactions of multiple clinical events. And capturing temporal interactions in the long event sequence is hard for traditional LSTM. Rather than directly applying the LSTM model to the event sequences, our proposed model firstly aggregates heterogeneous clinical events in a short period and then captures temporal interactions of the aggregated representations with LSTM. Our proposed Heterogeneous Event Aggregation can not only shorten the length of clinical event sequence but also help to retain temporal interactions of both categorical and numerical features of clinical events in the multiple heads of the aggregation representations. In the PhysioNet/Computing in Cardiology Challenge 2019, with the team named PKU_DLIB, our proposed model, in high efficiency, achieved utility score (0.321) in the full test set.
Learning Hierarchical Representations of Electronic Health Records for Clinical Outcome Prediction
Mar 20, 2019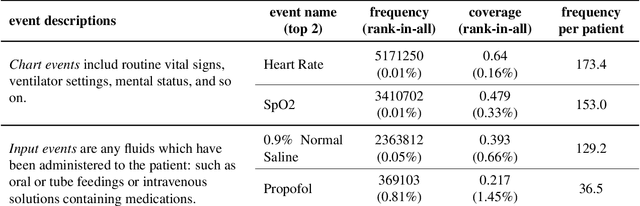
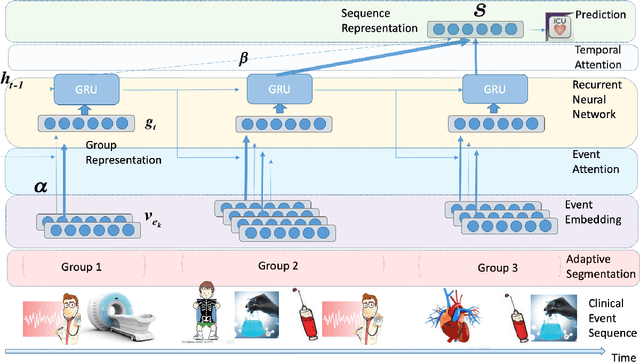
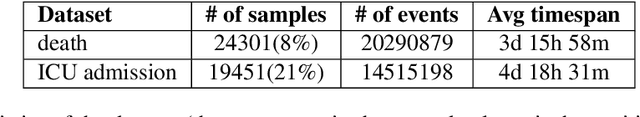
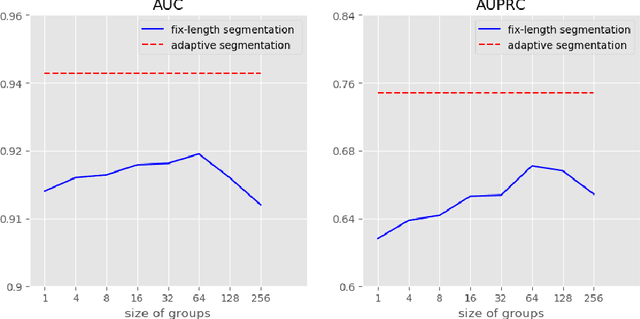
Abstract:Clinical outcome prediction based on the Electronic Health Record (EHR) plays a crucial role in improving the quality of healthcare. Conventional deep sequential models fail to capture the rich temporal patterns encoded in the longand irregular clinical event sequences. We make the observation that clinical events at a long time scale exhibit strongtemporal patterns, while events within a short time period tend to be disordered co-occurrence. We thus propose differentiated mechanisms to model clinical events at different time scales. Our model learns hierarchical representationsof event sequences, to adaptively distinguish between short-range and long-range events, and accurately capture coretemporal dependencies. Experimental results on real clinical data show that our model greatly improves over previous state-of-the-art models, achieving AUC scores of 0.94 and 0.90 for predicting death and ICU admission respectively, Our model also successfully identifies important events for different clinical outcome prediction tasks
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge