Zhihui Guo
MMEvalPro: Calibrating Multimodal Benchmarks Towards Trustworthy and Efficient Evaluation
Jun 29, 2024



Abstract:Large Multimodal Models (LMMs) exhibit impressive cross-modal understanding and reasoning abilities, often assessed through multiple-choice questions (MCQs) that include an image, a question, and several options. However, many benchmarks used for such evaluations suffer from systematic biases. Remarkably, Large Language Models (LLMs) without any visual perception capabilities achieve non-trivial performance, undermining the credibility of these evaluations. To address this issue while maintaining the efficiency of MCQ evaluations, we propose MMEvalPro, a benchmark designed to avoid Type-I errors through a trilogy evaluation pipeline and more rigorous metrics. For each original question from existing benchmarks, human annotators augment it by creating one perception question and one knowledge anchor question through a meticulous annotation process. MMEvalPro comprises $2,138$ question triplets, totaling $6,414$ distinct questions. Two-thirds of these questions are manually labeled by human experts, while the rest are sourced from existing benchmarks (MMMU, ScienceQA, and MathVista). Compared with the existing benchmarks, our experiments with the latest LLMs and LMMs demonstrate that MMEvalPro is more challenging (the best LMM lags behind human performance by $31.73\%$, compared to an average gap of $8.03\%$ in previous benchmarks) and more trustworthy (the best LLM trails the best LMM by $23.09\%$, whereas the gap for previous benchmarks is just $14.64\%$). Our in-depth analysis explains the reason for the large performance gap and justifies the trustworthiness of evaluation, underscoring its significant potential for advancing future research.
MM-Deacon: Multimodal molecular domain embedding analysis via contrastive learning
Sep 18, 2021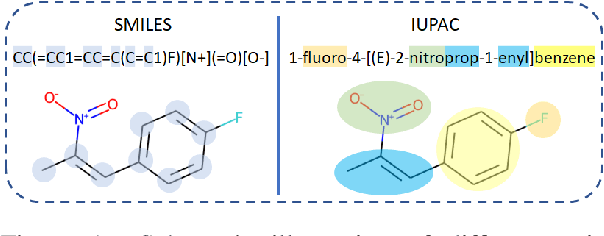
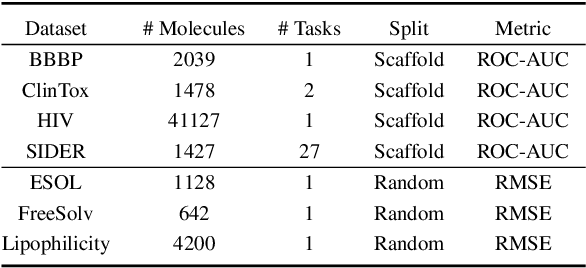
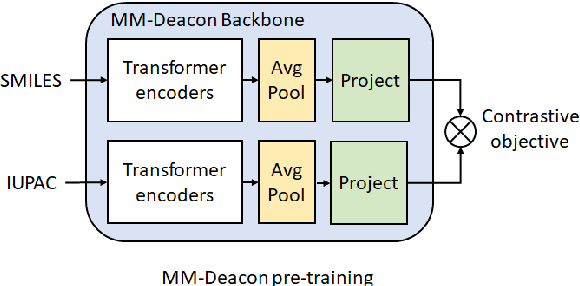
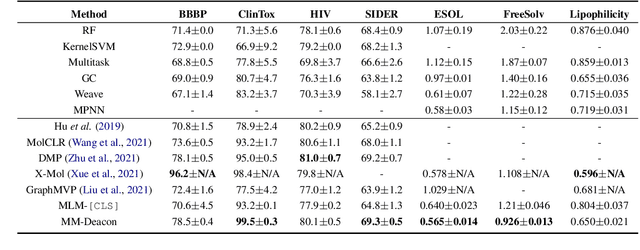
Abstract:Molecular representation learning plays an essential role in cheminformatics. Recently, language model-based approaches have been popular as an alternative to traditional expert-designed features to encode molecules. However, these approaches only utilize a single modality for representing molecules. Driven by the fact that a given molecule can be described through different modalities such as Simplified Molecular Line Entry System (SMILES), The International Union of Pure and Applied Chemistry (IUPAC), and The IUPAC International Chemical Identifier (InChI), we propose a multimodal molecular embedding generation approach called MM-Deacon (multimodal molecular domain embedding analysis via contrastive learning). MM-Deacon is trained using SMILES and IUPAC molecule representations as two different modalities. First, SMILES and IUPAC strings are encoded by using two different transformer-based language models independently, then the contrastive loss is utilized to bring these encoded representations from different modalities closer to each other if they belong to the same molecule, and to push embeddings farther from each other if they belong to different molecules. We evaluate the robustness of our molecule embeddings on molecule clustering, cross-modal molecule search, drug similarity assessment and drug-drug interaction tasks.
FilterNet: A Neighborhood Relationship Enhanced Fully Convolutional Network for Calf Muscle Compartment Segmentation
Jun 21, 2020



Abstract:Automated segmentation of individual calf muscle compartments from 3D magnetic resonance (MR) images is essential for developing quantitative biomarkers for muscular disease progression and its prediction. Achieving clinically acceptable results is a challenging task due to large variations in muscle shape and MR appearance. Although deep convolutional neural networks (DCNNs) achieved improved accuracy in various image segmentation tasks, certain problems such as utilizing long-range information and incorporating high-level constraints remain unsolved. We present a novel fully convolutional network (FCN), called FilterNet, that utilizes contextual information in a large neighborhood and embeds edge-aware constraints for individual calf muscle compartment segmentations. An encoder-decoder architecture with flexible backbone blocks is used to systematically enlarge convolution receptive field and preserve information at all resolutions. Edge positions derived from the FCN output muscle probability maps are explicitly regularized using kernel-based edge detection in an end-to-end optimization framework. Our FilterNet was evaluated on 40 T1-weighted MR images of 10 healthy and 30 diseased subjects by 4-fold cross-validation. Mean DICE coefficients of 88.00%--91.29% and mean absolute surface positioning errors of 1.04--1.66 mm were achieved for the five 3D muscle compartments.
DeepCenterline: a Multi-task Fully Convolutional Network for Centerline Extraction
Mar 25, 2019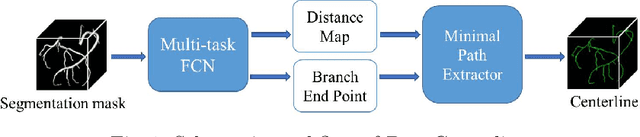

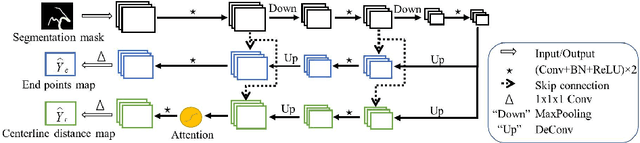
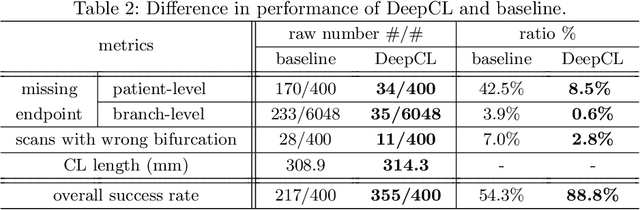
Abstract:A novel centerline extraction framework is reported which combines an end-to-end trainable multi-task fully convolutional network (FCN) with a minimal path extractor. The FCN simultaneously computes centerline distance maps and detects branch endpoints. The method generates single-pixel-wide centerlines with no spurious branches. It handles arbitrary tree-structured object with no prior assumption regarding depth of the tree or its bifurcation pattern. It is also robust to substantial scale changes across different parts of the target object and minor imperfections of the object's segmentation mask. To the best of our knowledge, this is the first deep-learning based centerline extraction method that guarantees single-pixel-wide centerline for a complex tree-structured object. The proposed method is validated in coronary artery centerline extraction on a dataset of 620 patients (400 of which used as test set). This application is challenging due to the large number of coronary branches, branch tortuosity, and large variations in length, thickness, shape, etc. The proposed method generates well-positioned centerlines, exhibiting lower number of missing branches and is more robust in the presence of minor imperfections of the object segmentation mask. Compared to a state-of-the-art traditional minimal path approach, our method improves patient-level success rate of centerline extraction from 54.3% to 88.8% according to independent human expert review.
Deep LOGISMOS: Deep Learning Graph-based 3D Segmentation of Pancreatic Tumors on CT scans
Jan 25, 2018



Abstract:This paper reports Deep LOGISMOS approach to 3D tumor segmentation by incorporating boundary information derived from deep contextual learning to LOGISMOS - layered optimal graph image segmentation of multiple objects and surfaces. Accurate and reliable tumor segmentation is essential to tumor growth analysis and treatment selection. A fully convolutional network (FCN), UNet, is first trained using three adjacent 2D patches centered at the tumor, providing contextual UNet segmentation and probability map for each 2D patch. The UNet segmentation is then refined by Gaussian Mixture Model (GMM) and morphological operations. The refined UNet segmentation is used to provide the initial shape boundary to build a segmentation graph. The cost for each node of the graph is determined by the UNet probability maps. Finally, a max-flow algorithm is employed to find the globally optimal solution thus obtaining the final segmentation. For evaluation, we applied the method to pancreatic tumor segmentation on a dataset of 51 CT scans, among which 30 scans were used for training and 21 for testing. With Deep LOGISMOS, DICE Similarity Coefficient (DSC) and Relative Volume Difference (RVD) reached 83.2+-7.8% and 18.6+-17.4% respectively, both are significantly improved (p<0.05) compared with contextual UNet and/or LOGISMOS alone.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge