Robin Abraham
BatchPrompt: Accomplish more with less
Sep 05, 2023



Abstract:As the ever-increasing token limits of large language models (LLMs) have enabled long context as input, prompting with single data samples might no longer an efficient way. A straightforward strategy improving efficiency is to batch data within the token limit (e.g., 8k for gpt-3.5-turbo; 32k for GPT-4), which we call BatchPrompt. We have two initial observations for prompting with batched data. First, we find that prompting with batched data in longer contexts will inevitably lead to worse performance, compared to single-data prompting. Second, the performance of the language model is significantly correlated with the positions and order of the batched data, due to the corresponding change in decoder context. To retain efficiency and overcome performance loss, we propose Batch Permutation and Ensembling (BPE), and a novel Self-reflection-guided EArly Stopping (SEAS) technique. Our comprehensive experimental evaluation demonstrates that BPE can boost the performance of BatchPrompt with a striking margin on a range of popular NLP tasks, including question answering (Boolq), textual entailment (RTE), and duplicate questions identification (QQP). These performances are even competitive with/higher than single-data prompting(SinglePrompt), while BatchPrompt requires much fewer LLM calls and input tokens (For SinglePrompt v.s. BatchPrompt with batch size 32, using just 9%-16% the number of LLM calls, Boolq accuracy 90.6% to 90.9% with 27.4% tokens, QQP accuracy 87.2% to 88.4% with 18.6% tokens, RTE accuracy 91.5% to 91.1% with 30.8% tokens). To the best of our knowledge, this is the first work to technically improve prompting efficiency of large language models. We hope our simple yet effective approach will shed light on the future research of large language models. The code will be released.
Aligning benchmark datasets for table structure recognition
Mar 01, 2023Abstract:Benchmark datasets for table structure recognition (TSR) must be carefully processed to ensure they are annotated consistently. However, even if a dataset's annotations are self-consistent, there may be significant inconsistency across datasets, which can harm the performance of models trained and evaluated on them. In this work, we show that aligning these benchmarks$\unicode{x2014}$removing both errors and inconsistency between them$\unicode{x2014}$improves model performance significantly. We demonstrate this through a data-centric approach where we adopt a single model architecture, the Table Transformer (TATR), that we hold fixed throughout. Baseline exact match accuracy for TATR evaluated on the ICDAR-2013 benchmark is 65% when trained on PubTables-1M, 42% when trained on FinTabNet, and 69% combined. After reducing annotation mistakes and inter-dataset inconsistency, performance of TATR evaluated on ICDAR-2013 increases substantially to 75% when trained on PubTables-1M, 65% when trained on FinTabNet, and 81% combined. We show through ablations over the modification steps that canonicalization of the table annotations has a significantly positive effect on performance, while other choices balance necessary trade-offs that arise when deciding a benchmark dataset's final composition. Overall we believe our work has significant implications for benchmark design for TSR and potentially other tasks as well. All dataset processing and training code will be released.
uGLAD: Sparse graph recovery by optimizing deep unrolled networks
May 23, 2022
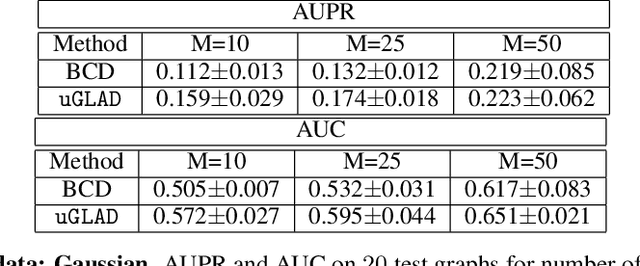
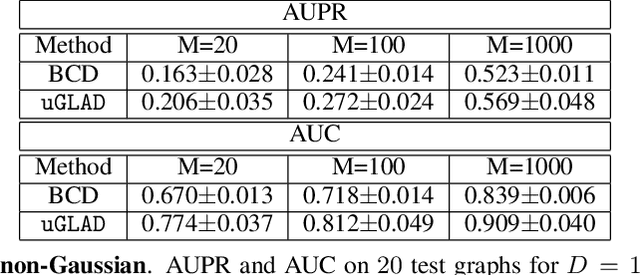
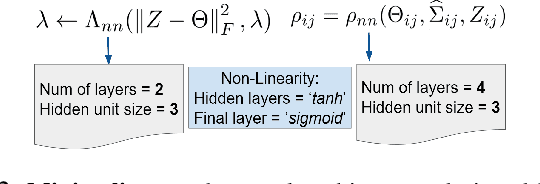
Abstract:Probabilistic Graphical Models (PGMs) are generative models of complex systems. They rely on conditional independence assumptions between variables to learn sparse representations which can be visualized in a form of a graph. Such models are used for domain exploration and structure discovery in poorly understood domains. This work introduces a novel technique to perform sparse graph recovery by optimizing deep unrolled networks. Assuming that the input data $X\in\mathbb{R}^{M\times D}$ comes from an underlying multivariate Gaussian distribution, we apply a deep model on $X$ that outputs the precision matrix $\Theta$, which can also be interpreted as the adjacency matrix. Our model, uGLAD, builds upon and extends the state-of-the-art model GLAD to the unsupervised setting. The key benefits of our model are (1) uGLAD automatically optimizes sparsity-related regularization parameters leading to better performance than existing algorithms. (2) We introduce multi-task learning based `consensus' strategy for robust handling of missing data in an unsupervised setting. We evaluate model results on synthetic Gaussian data, non-Gaussian data generated from Gene Regulatory Networks, and present a case study in anaerobic digestion.
GriTS: Grid table similarity metric for table structure recognition
Mar 23, 2022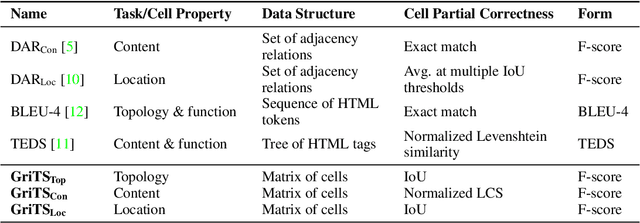
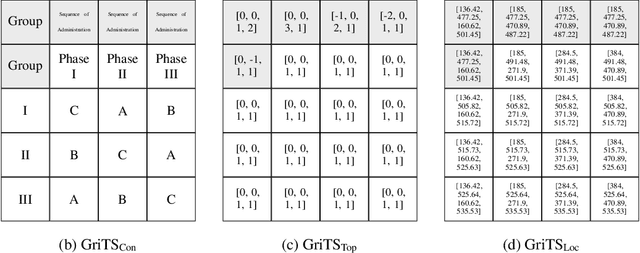
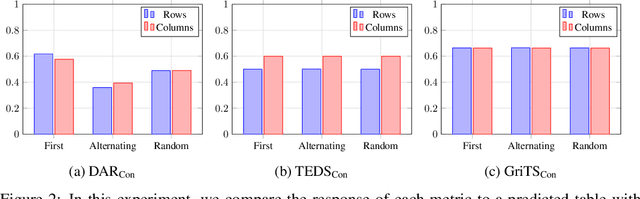
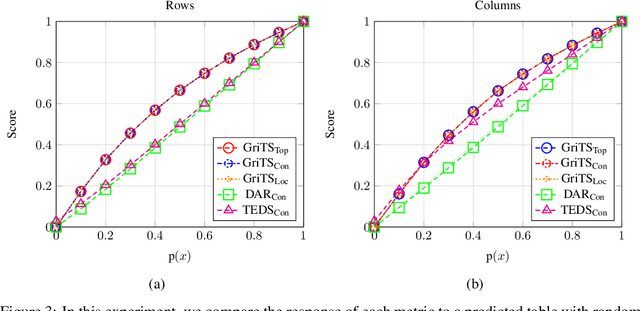
Abstract:In this paper, we propose a new class of evaluation metric for table structure recognition, grid table similarity (GriTS). Unlike prior metrics, GriTS evaluates the correctness of a predicted table directly in its natural form as a matrix. To create a similarity measure between matrices, we generalize the two-dimensional largest common substructure (2D-LCS) problem, which is NP-hard, to the 2D most similar substructures (2D-MSS) problem and propose a polynomial-time heuristic for solving it. We validate empirically using the PubTables-1M dataset that comparison between matrices exhibits more desirable behavior than alternatives for table structure recognition evaluation. GriTS also unifies all three subtasks of cell topology recognition, cell location recognition, and cell content recognition within the same framework, which simplifies the evaluation and enables more meaningful comparisons across different types of structure recognition approaches. Code will be released at https://github.com/microsoft/table-transformer.
PubTables-1M: Towards a universal dataset and metrics for training and evaluating table extraction models
Oct 12, 2021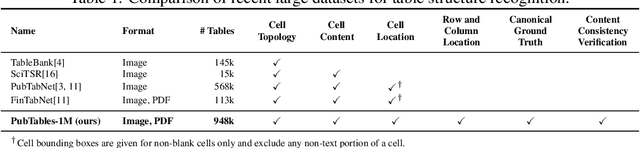
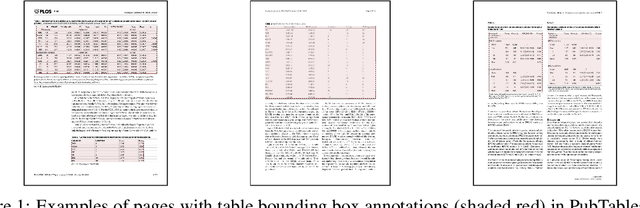

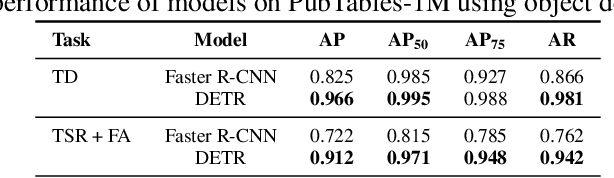
Abstract:Recently, interest has grown in applying machine learning to the problem of table structure inference and extraction from unstructured documents. However, progress in this area has been challenging both to make and to measure, due to several issues that arise in training and evaluating models from labeled data. This includes challenges as fundamental as the lack of a single definitive ground truth output for each input sample and the lack of an ideal metric for measuring partial correctness for this task. To address these issues we propose a new dataset, PubMed Tables One Million (PubTables-1M), and a new class of metric, grid table similarity (GriTS). PubTables-1M is nearly twice as large as the previous largest comparable dataset, contains highly-detailed structure annotations, and can be used for models across multiple architectures and modalities. Further, it addresses issues such as ambiguity and lack of consistency in the annotations via a novel canonicalization and quality control procedure. We apply DETR to table extraction for the first time and show that object detection models trained on PubTables-1M produce excellent results out-of-the-box for all three tasks of detection, structure recognition, and functional analysis. It is our hope that PubTables-1M and GriTS can further progress in this area by creating data and metrics suitable for training and evaluating a wide variety of models for table extraction. Data and code will be released at https://github.com/microsoft/table-transformer.
MM-Deacon: Multimodal molecular domain embedding analysis via contrastive learning
Sep 18, 2021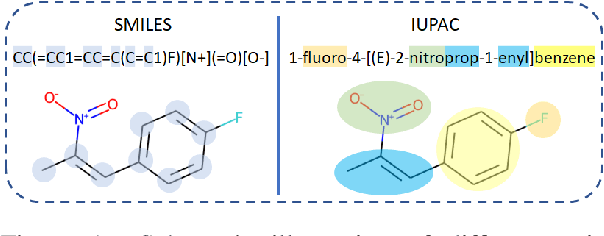
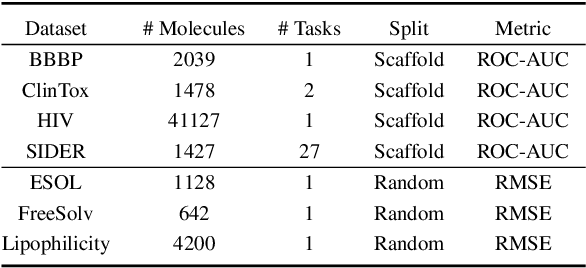
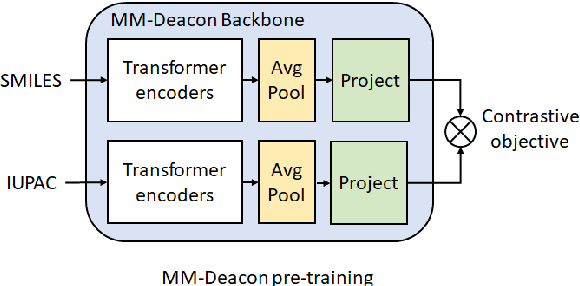
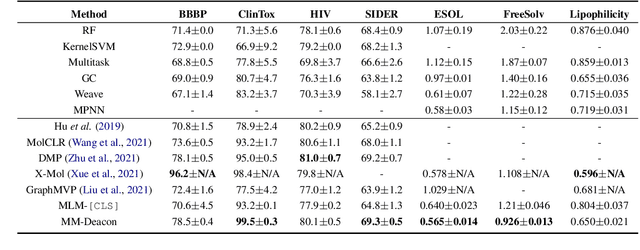
Abstract:Molecular representation learning plays an essential role in cheminformatics. Recently, language model-based approaches have been popular as an alternative to traditional expert-designed features to encode molecules. However, these approaches only utilize a single modality for representing molecules. Driven by the fact that a given molecule can be described through different modalities such as Simplified Molecular Line Entry System (SMILES), The International Union of Pure and Applied Chemistry (IUPAC), and The IUPAC International Chemical Identifier (InChI), we propose a multimodal molecular embedding generation approach called MM-Deacon (multimodal molecular domain embedding analysis via contrastive learning). MM-Deacon is trained using SMILES and IUPAC molecule representations as two different modalities. First, SMILES and IUPAC strings are encoded by using two different transformer-based language models independently, then the contrastive loss is utilized to bring these encoded representations from different modalities closer to each other if they belong to the same molecule, and to push embeddings farther from each other if they belong to different molecules. We evaluate the robustness of our molecule embeddings on molecule clustering, cross-modal molecule search, drug similarity assessment and drug-drug interaction tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge