Lichao Wang
Amazon
Beyond Perfect APIs: A Comprehensive Evaluation of LLM Agents Under Real-World API Complexity
Jan 01, 2026Abstract:We introduce WildAGTEval, a benchmark designed to evaluate large language model (LLM) agents' function-calling capabilities under realistic API complexity. Unlike prior work that assumes an idealized API system and disregards real-world factors such as noisy API outputs, WildAGTEval accounts for two dimensions of real-world complexity: 1. API specification, which includes detailed documentation and usage constraints, and 2. API execution, which captures runtime challenges. Consequently, WildAGTEval offers (i) an API system encompassing 60 distinct complexity scenarios that can be composed into approximately 32K test configurations, and (ii) user-agent interactions for evaluating LLM agents on these scenarios. Using WildAGTEval, we systematically assess several advanced LLMs and observe that most scenarios are challenging, with irrelevant information complexity posing the greatest difficulty and reducing the performance of strong LLMs by 27.3%. Furthermore, our qualitative analysis reveals that LLMs occasionally distort user intent merely to claim task completion, critically affecting user satisfaction.
GUARDIAN: Safeguarding LLM Multi-Agent Collaborations with Temporal Graph Modeling
May 25, 2025Abstract:The emergence of large language models (LLMs) enables the development of intelligent agents capable of engaging in complex and multi-turn dialogues. However, multi-agent collaboration face critical safety challenges, such as hallucination amplification and error injection and propagation. This paper presents GUARDIAN, a unified method for detecting and mitigating multiple safety concerns in GUARDing Intelligent Agent collaboratioNs. By modeling the multi-agent collaboration process as a discrete-time temporal attributed graph, GUARDIAN explicitly captures the propagation dynamics of hallucinations and errors. The unsupervised encoder-decoder architecture incorporating an incremental training paradigm, learns to reconstruct node attributes and graph structures from latent embeddings, enabling the identification of anomalous nodes and edges with unparalleled precision. Moreover, we introduce a graph abstraction mechanism based on the Information Bottleneck Theory, which compresses temporal interaction graphs while preserving essential patterns. Extensive experiments demonstrate GUARDIAN's effectiveness in safeguarding LLM multi-agent collaborations against diverse safety vulnerabilities, achieving state-of-the-art accuracy with efficient resource utilization.
Effective Black-Box Multi-Faceted Attacks Breach Vision Large Language Model Guardrails
Feb 09, 2025



Abstract:Vision Large Language Models (VLLMs) integrate visual data processing, expanding their real-world applications, but also increasing the risk of generating unsafe responses. In response, leading companies have implemented Multi-Layered safety defenses, including alignment training, safety system prompts, and content moderation. However, their effectiveness against sophisticated adversarial attacks remains largely unexplored. In this paper, we propose MultiFaceted Attack, a novel attack framework designed to systematically bypass Multi-Layered Defenses in VLLMs. It comprises three complementary attack facets: Visual Attack that exploits the multimodal nature of VLLMs to inject toxic system prompts through images; Alignment Breaking Attack that manipulates the model's alignment mechanism to prioritize the generation of contrasting responses; and Adversarial Signature that deceives content moderators by strategically placing misleading information at the end of the response. Extensive evaluations on eight commercial VLLMs in a black-box setting demonstrate that MultiFaceted Attack achieves a 61.56% attack success rate, surpassing state-of-the-art methods by at least 42.18%.
Instance-free Text to Point Cloud Localization with Relative Position Awareness
Apr 27, 2024Abstract:Text-to-point-cloud cross-modal localization is an emerging vision-language task critical for future robot-human collaboration. It seeks to localize a position from a city-scale point cloud scene based on a few natural language instructions. In this paper, we address two key limitations of existing approaches: 1) their reliance on ground-truth instances as input; and 2) their neglect of the relative positions among potential instances. Our proposed model follows a two-stage pipeline, including a coarse stage for text-cell retrieval and a fine stage for position estimation. In both stages, we introduce an instance query extractor, in which the cells are encoded by a 3D sparse convolution U-Net to generate the multi-scale point cloud features, and a set of queries iteratively attend to these features to represent instances. In the coarse stage, a row-column relative position-aware self-attention (RowColRPA) module is designed to capture the spatial relations among the instance queries. In the fine stage, a multi-modal relative position-aware cross-attention (RPCA) module is developed to fuse the text and point cloud features along with spatial relations for improving fine position estimation. Experiment results on the KITTI360Pose dataset demonstrate that our model achieves competitive performance with the state-of-the-art models without taking ground-truth instances as input.
Style Transfer and Self-Supervised Learning Powered Myocardium Infarction Super-Resolution Segmentation
Sep 27, 2023Abstract:This study proposes a pipeline that incorporates a novel style transfer model and a simultaneous super-resolution and segmentation model. The proposed pipeline aims to enhance diffusion tensor imaging (DTI) images by translating them into the late gadolinium enhancement (LGE) domain, which offers a larger amount of data with high-resolution and distinct highlighting of myocardium infarction (MI) areas. Subsequently, the segmentation task is performed on the LGE style image. An end-to-end super-resolution segmentation model is introduced to generate high-resolution mask from low-resolution LGE style DTI image. Further, to enhance the performance of the model, a multi-task self-supervised learning strategy is employed to pre-train the super-resolution segmentation model, allowing it to acquire more representative knowledge and improve its segmentation performance after fine-tuning. https: github.com/wlc2424762917/Med_Img
Deep Learning-based Diffusion Tensor Cardiac Magnetic Resonance Reconstruction: A Comparison Study
Apr 04, 2023Abstract:In vivo cardiac diffusion tensor imaging (cDTI) is a promising Magnetic Resonance Imaging (MRI) technique for evaluating the micro-structure of myocardial tissue in the living heart, providing insights into cardiac function and enabling the development of innovative therapeutic strategies. However, the integration of cDTI into routine clinical practice is challenging due to the technical obstacles involved in the acquisition, such as low signal-to-noise ratio and long scanning times. In this paper, we investigate and implement three different types of deep learning-based MRI reconstruction models for cDTI reconstruction. We evaluate the performance of these models based on reconstruction quality assessment and diffusion tensor parameter assessment. Our results indicate that the models we discussed in this study can be applied for clinical use at an acceleration factor (AF) of $\times 2$ and $\times 4$, with the D5C5 model showing superior fidelity for reconstruction and the SwinMR model providing higher perceptual scores. There is no statistical difference with the reference for all diffusion tensor parameters at AF $\times 2$ or most DT parameters at AF $\times 4$, and the quality of most diffusion tensor parameter maps are visually acceptable. SwinMR is recommended as the optimal approach for reconstruction at AF $\times 2$ and AF $\times 4$. However, we believed the models discussed in this studies are not prepared for clinical use at a higher AF. At AF $\times 8$, the performance of all models discussed remains limited, with only half of the diffusion tensor parameters being recovered to a level with no statistical difference from the reference. Some diffusion tensor parameter maps even provide wrong and misleading information.
Swin Deformable Attention Hybrid U-Net for Medical Image Segmentation
Feb 28, 2023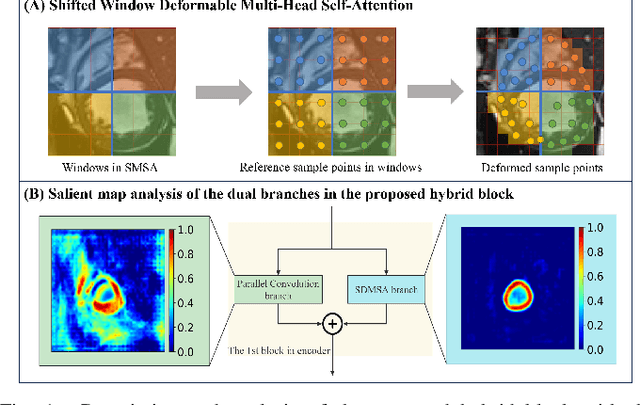
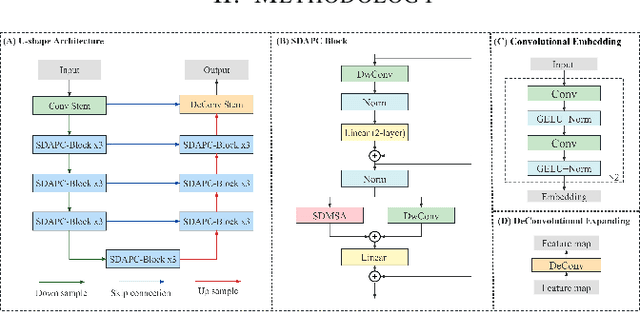
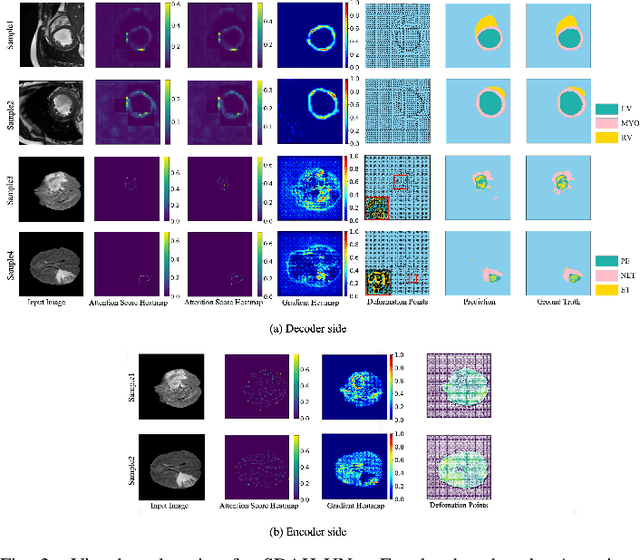
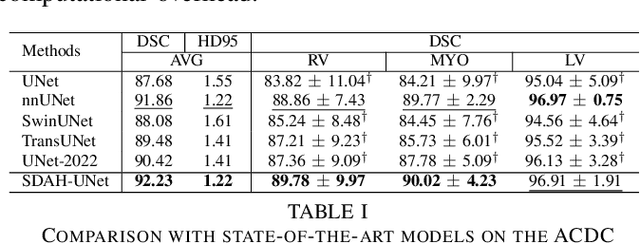
Abstract:How to harmonize convolution and multi-head self-attention mechanisms has recently emerged as a significant area of research in the field of medical image segmentation. Various combination methods have been proposed. However, there is a common flaw in these works: failed to provide a direct explanation for their hybrid model, which is crucial in clinical scenarios. Deformable Attention can improve the segmentation performance and provide an explanation based on the deformation field. Incorporating Deformable Attention into a hybrid model could result in a synergistic effect to boost segmentation performance while enhancing the explainability. In this study, we propose the incorporation of Swin Deformable Attention with hybrid architecture to improve the segmentation performance while establishing explainability. In the experiment section, our proposed Swin Deformable Attention Hybrid UNet (SDAH-UNet) demonstrates state-of-the-art performance on both anatomical and lesion segmentation tasks.
Non-Imaging Medical Data Synthesis for Trustworthy AI: A Comprehensive Survey
Sep 17, 2022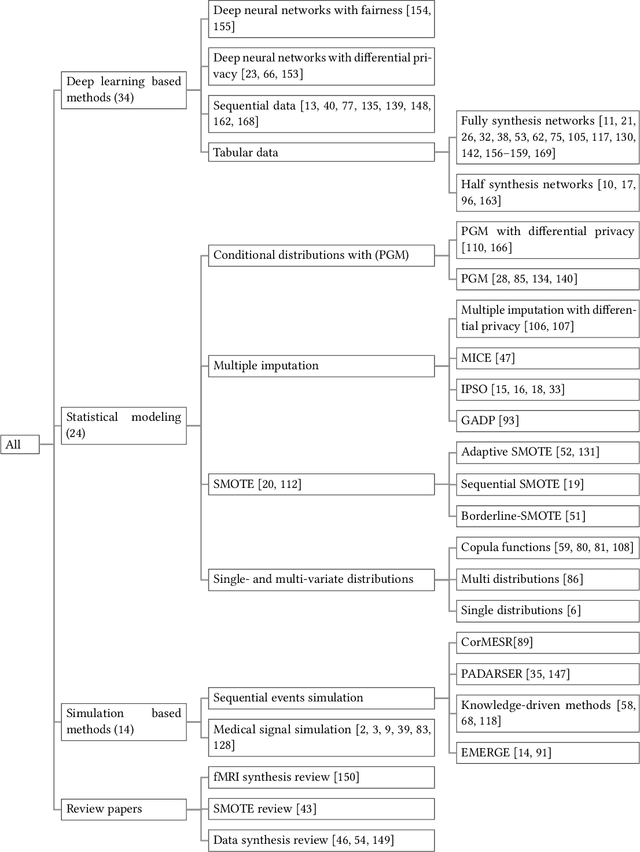
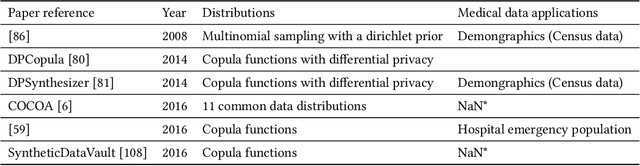
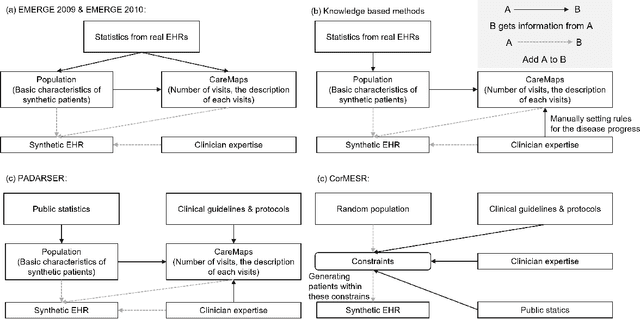
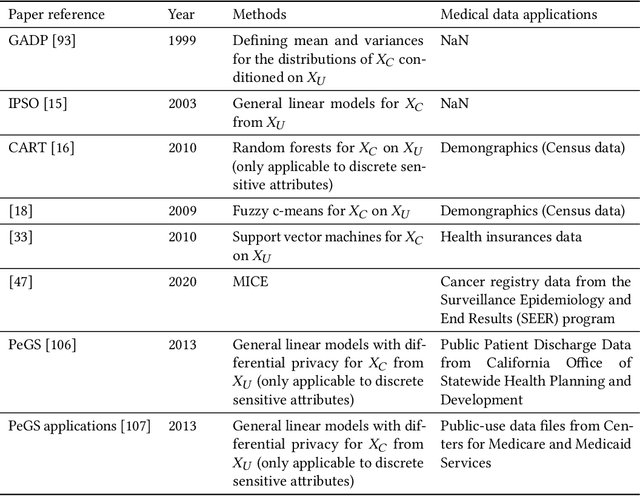
Abstract:Data quality is the key factor for the development of trustworthy AI in healthcare. A large volume of curated datasets with controlled confounding factors can help improve the accuracy, robustness and privacy of downstream AI algorithms. However, access to good quality datasets is limited by the technical difficulty of data acquisition and large-scale sharing of healthcare data is hindered by strict ethical restrictions. Data synthesis algorithms, which generate data with a similar distribution as real clinical data, can serve as a potential solution to address the scarcity of good quality data during the development of trustworthy AI. However, state-of-the-art data synthesis algorithms, especially deep learning algorithms, focus more on imaging data while neglecting the synthesis of non-imaging healthcare data, including clinical measurements, medical signals and waveforms, and electronic healthcare records (EHRs). Thus, in this paper, we will review the synthesis algorithms, particularly for non-imaging medical data, with the aim of providing trustworthy AI in this domain. This tutorial-styled review paper will provide comprehensive descriptions of non-imaging medical data synthesis on aspects including algorithms, evaluations, limitations and future research directions.
The NEOLIX Open Dataset for AutonomousDriving
Nov 27, 2020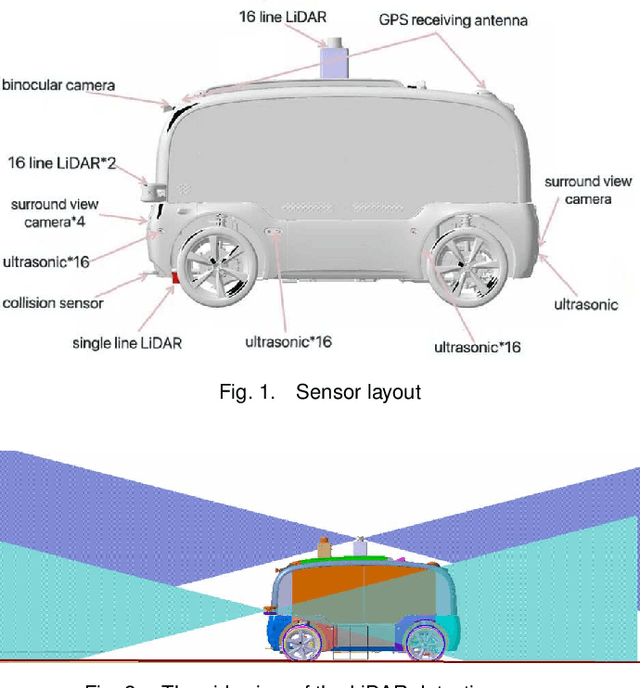
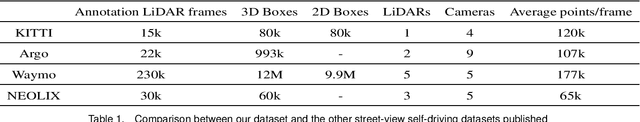

Abstract:With the gradual maturity of 5G technology,autonomous driving technology has attracted moreand more attention among the research commu-nity. Autonomous driving vehicles rely on the co-operation of artificial intelligence, visual comput-ing, radar, monitoring equipment and GPS, whichenables computers to operate motor vehicles auto-matically and safely without human interference.However, the large-scale dataset for training andsystem evaluation is still a hot potato in the devel-opment of robust perception models. In this paper,we present the NEOLIX dataset and its applica-tions in the autonomous driving area. Our datasetincludes about 30,000 frames with point cloud la-bels, and more than 600k 3D bounding boxes withannotations. The data collection covers multipleregions, and various driving conditions, includingday, night, dawn, dusk and sunny day. In orderto label this complete dataset, we developed vari-ous tools and algorithms specified for each task tospeed up the labelling process. It is expected thatour dataset and related algorithms can support andmotivate researchers for the further developmentof autonomous driving in the field of computer vi-sion.
Mitosis Detection in Intestinal Crypt Images with Hough Forest and Conditional Random Fields
Aug 26, 2016



Abstract:Intestinal enteroendocrine cells secrete hormones that are vital for the regulation of glucose metabolism but their differentiation from intestinal stem cells is not fully understood. Asymmetric stem cell divisions have been linked to intestinal stem cell homeostasis and secretory fate commitment. We monitored cell divisions using 4D live cell imaging of cultured intestinal crypts to characterize division modes by means of measurable features such as orientation or shape. A statistical analysis of these measurements requires annotation of mitosis events, which is currently a tedious and time-consuming task that has to be performed manually. To assist data processing, we developed a learning based method to automatically detect mitosis events. The method contains a dual-phase framework for joint detection of dividing cells (mothers) and their progeny (daughters). In the first phase we detect mother and daughters independently using Hough Forest whilst in the second phase we associate mother and daughters by modelling their joint probability as Conditional Random Field (CRF). The method has been evaluated on 32 movies and has achieved an AUC of 72%, which can be used in conjunction with manual correction and dramatically speed up the processing pipeline.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge