Jingru Yi
Augment the Pairs: Semantics-Preserving Image-Caption Pair Augmentation for Grounding-Based Vision and Language Models
Nov 05, 2023Abstract:Grounding-based vision and language models have been successfully applied to low-level vision tasks, aiming to precisely locate objects referred in captions. The effectiveness of grounding representation learning heavily relies on the scale of the training dataset. Despite being a useful data enrichment strategy, data augmentation has received minimal attention in existing vision and language tasks as augmentation for image-caption pairs is non-trivial. In this study, we propose a robust phrase grounding model trained with text-conditioned and text-unconditioned data augmentations. Specifically, we apply text-conditioned color jittering and horizontal flipping to ensure semantic consistency between images and captions. To guarantee image-caption correspondence in the training samples, we modify the captions according to pre-defined keywords when applying horizontal flipping. Additionally, inspired by recent masked signal reconstruction, we propose to use pixel-level masking as a novel form of data augmentation. While we demonstrate our data augmentation method with MDETR framework, the proposed approach is applicable to common grounding-based vision and language tasks with other frameworks. Finally, we show that image encoder pretrained on large-scale image and language datasets (such as CLIP) can further improve the results. Through extensive experiments on three commonly applied datasets: Flickr30k, referring expressions and GQA, our method demonstrates advanced performance over the state-of-the-arts with various metrics. Code can be found in https://github.com/amzn/augment-the-pairs-wacv2024.
Region Proposal Rectification Towards Robust Instance Segmentation of Biological Images
Mar 06, 2022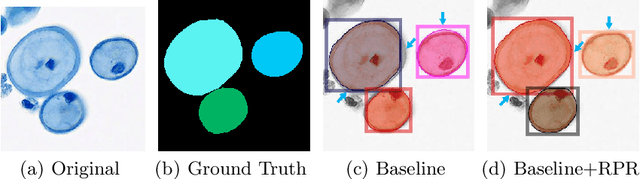
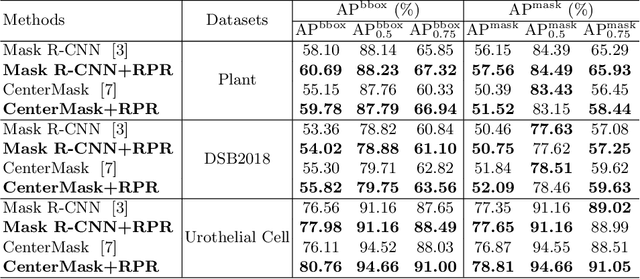
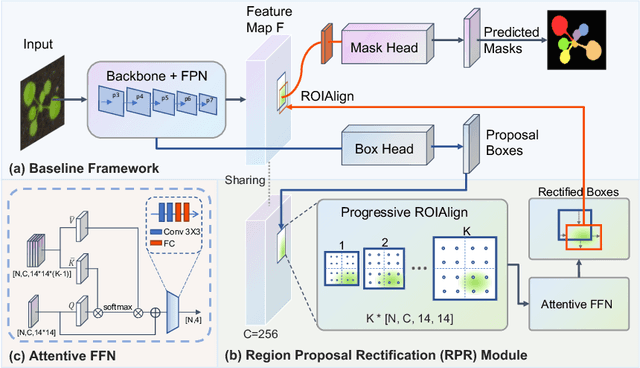
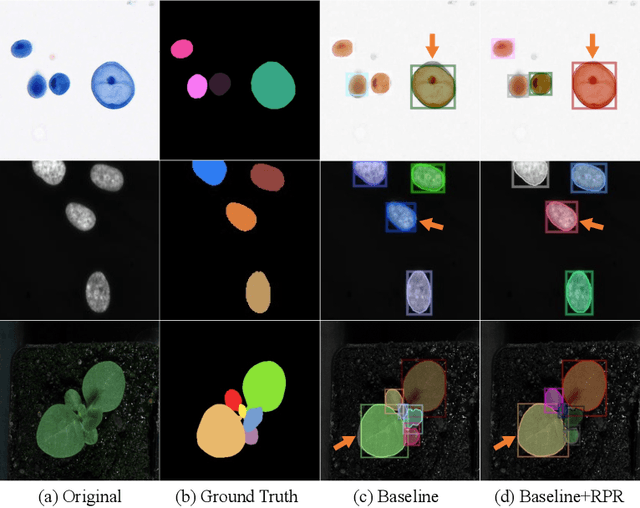
Abstract:Top-down instance segmentation framework has shown its superiority in object detection compared to the bottom-up framework. While it is efficient in addressing over-segmentation, top-down instance segmentation suffers from over-crop problem. However, a complete segmentation mask is crucial for biological image analysis as it delivers important morphological properties such as shapes and volumes. In this paper, we propose a region proposal rectification (RPR) module to address this challenging incomplete segmentation problem. In particular, we offer a progressive ROIAlign module to introduce neighbor information into a series of ROIs gradually. The ROI features are fed into an attentive feed-forward network (FFN) for proposal box regression. With additional neighbor information, the proposed RPR module shows significant improvement in correction of region proposal locations and thereby exhibits favorable instance segmentation performances on three biological image datasets compared to state-of-the-art baseline methods. Experimental results demonstrate that the proposed RPR module is effective in both anchor-based and anchor-free top-down instance segmentation approaches, suggesting the proposed method can be applied to general top-down instance segmentation of biological images.
Object-Guided Instance Segmentation With Auxiliary Feature Refinement for Biological Images
Jun 14, 2021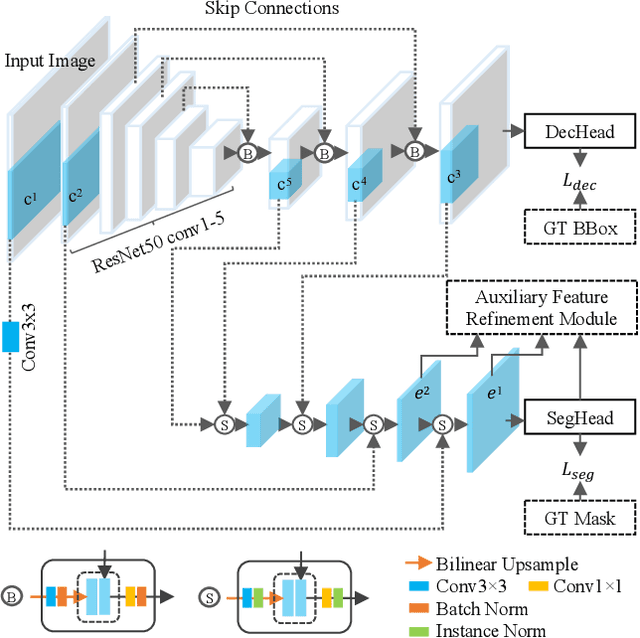
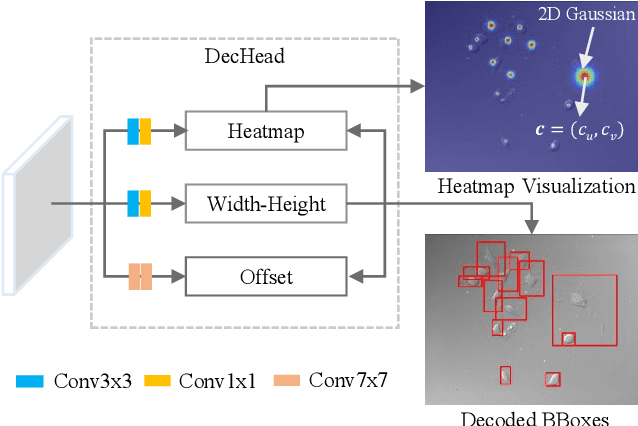
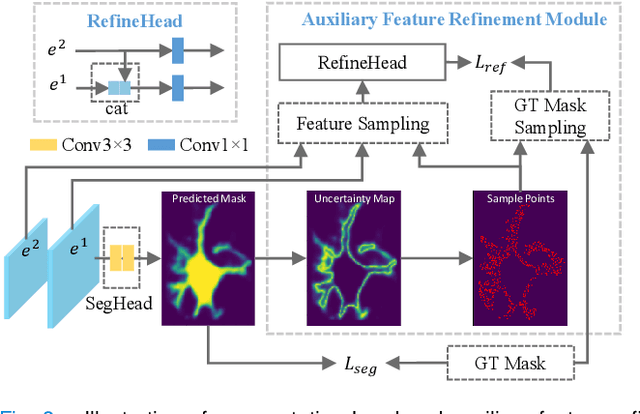
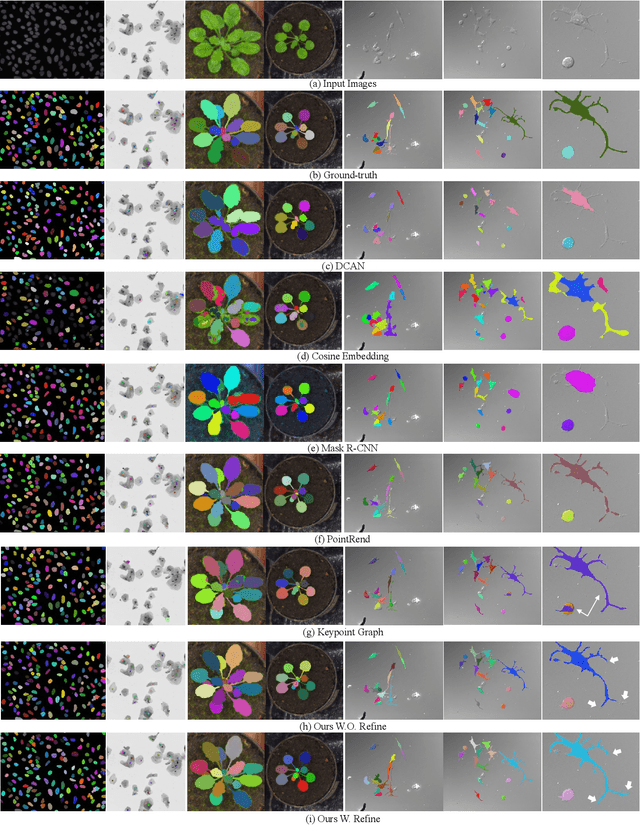
Abstract:Instance segmentation is of great importance for many biological applications, such as study of neural cell interactions, plant phenotyping, and quantitatively measuring how cells react to drug treatment. In this paper, we propose a novel box-based instance segmentation method. Box-based instance segmentation methods capture objects via bounding boxes and then perform individual segmentation within each bounding box region. However, existing methods can hardly differentiate the target from its neighboring objects within the same bounding box region due to their similar textures and low-contrast boundaries. To deal with this problem, in this paper, we propose an object-guided instance segmentation method. Our method first detects the center points of the objects, from which the bounding box parameters are then predicted. To perform segmentation, an object-guided coarse-to-fine segmentation branch is built along with the detection branch. The segmentation branch reuses the object features as guidance to separate target object from the neighboring ones within the same bounding box region. To further improve the segmentation quality, we design an auxiliary feature refinement module that densely samples and refines point-wise features in the boundary regions. Experimental results on three biological image datasets demonstrate the advantages of our method. The code will be available at https://github.com/yijingru/ObjGuided-Instance-Segmentation.
Oriented Object Detection in Aerial Images with Box Boundary-Aware Vectors
Aug 29, 2020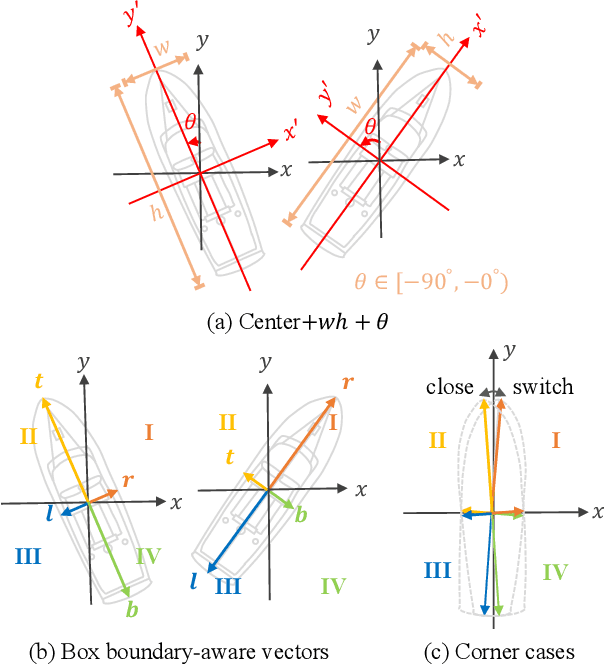
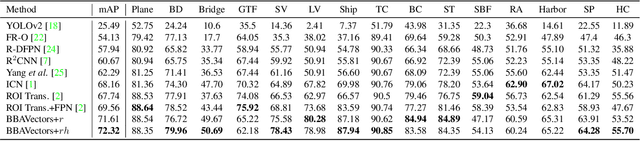
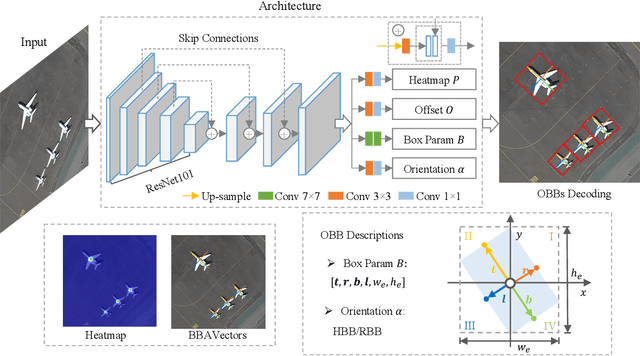
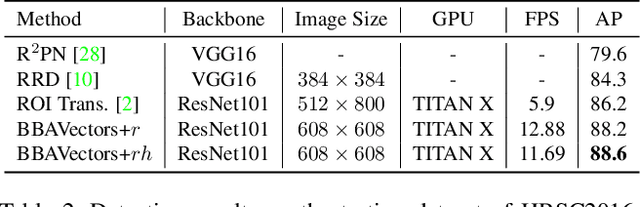
Abstract:Oriented object detection in aerial images is a challenging task as the objects in aerial images are displayed in arbitrary directions and are usually densely packed. Current oriented object detection methods mainly rely on two-stage anchor-based detectors. However, the anchor-based detectors typically suffer from a severe imbalance issue between the positive and negative anchor boxes. To address this issue, in this work we extend the horizontal keypoint-based object detector to the oriented object detection task. In particular, we first detect the center keypoints of the objects, based on which we then regress the box boundary-aware vectors (BBAVectors) to capture the oriented bounding boxes. The box boundary-aware vectors are distributed in the four quadrants of a Cartesian coordinate system for all arbitrarily oriented objects. To relieve the difficulty of learning the vectors in the corner cases, we further classify the oriented bounding boxes into horizontal and rotational bounding boxes. In the experiment, we show that learning the box boundary-aware vectors is superior to directly predicting the width, height, and angle of an oriented bounding box, as adopted in the baseline method. Besides, the proposed method competes favorably with state-of-the-art methods. Code is available at https://github.com/yijingru/BBAVectors-Oriented-Object-Detection.
Enhanced MRI Reconstruction Network using Neural Architecture Search
Aug 19, 2020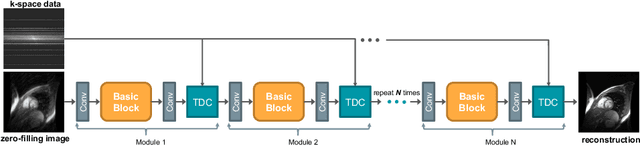
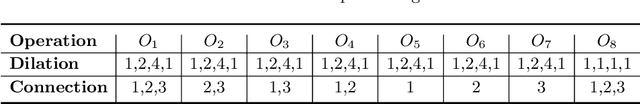
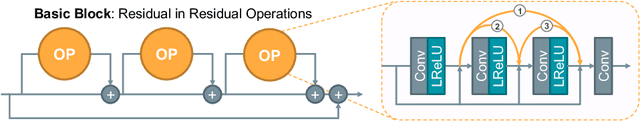
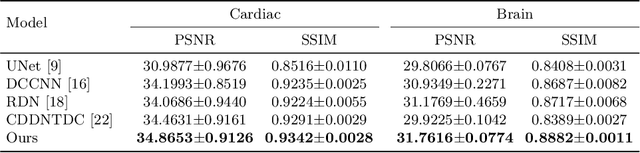
Abstract:The accurate reconstruction of under-sampled magnetic resonance imaging (MRI) data using modern deep learning technology, requires significant effort to design the necessary complex neural network architectures. The cascaded network architecture for MRI reconstruction has been widely used, while it suffers from the "vanishing gradient" problem when the network becomes deep. In addition, homogeneous architecture degrades the representation capacity of the network. In this work, we present an enhanced MRI reconstruction network using a residual in residual basic block. For each cell in the basic block, we use the differentiable neural architecture search (NAS) technique to automatically choose the optimal operation among eight variants of the dense block. This new heterogeneous network is evaluated on two publicly available datasets and outperforms all current state-of-the-art methods, which demonstrates the effectiveness of our proposed method.
PC-U Net: Learning to Jointly Reconstruct and Segment the Cardiac Walls in 3D from CT Data
Aug 18, 2020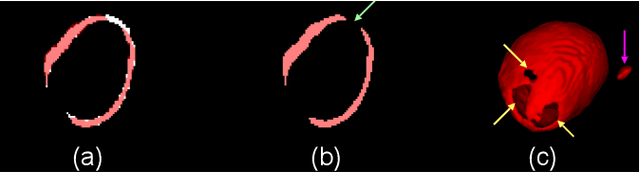
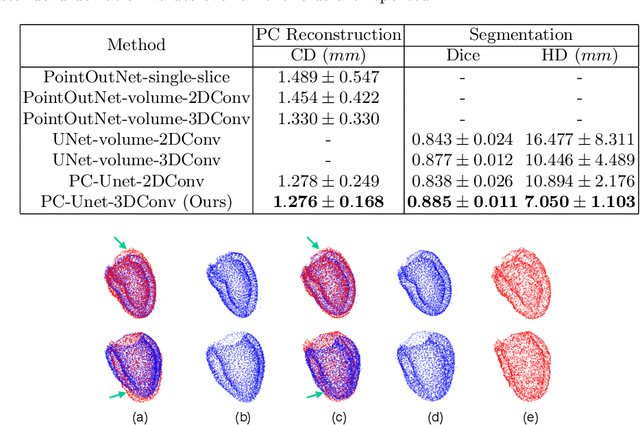
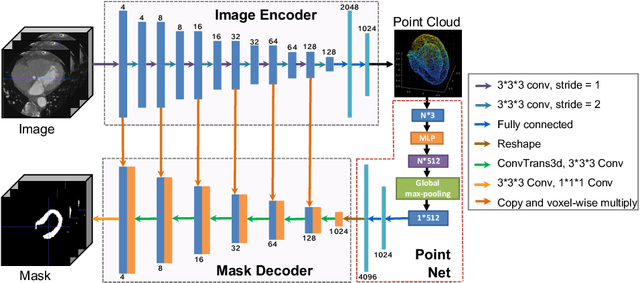
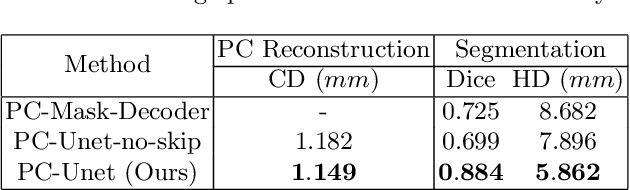
Abstract:The 3D volumetric shape of the heart's left ventricle (LV) myocardium (MYO) wall provides important information for diagnosis of cardiac disease and invasive procedure navigation. Many cardiac image segmentation methods have relied on detection of region-of-interest as a pre-requisite for shape segmentation and modeling. With segmentation results, a 3D surface mesh and a corresponding point cloud of the segmented cardiac volume can be reconstructed for further analyses. Although state-of-the-art methods (e.g., U-Net) have achieved decent performance on cardiac image segmentation in terms of accuracy, these segmentation results can still suffer from imaging artifacts and noise, which will lead to inaccurate shape modeling results. In this paper, we propose a PC-U net that jointly reconstructs the point cloud of the LV MYO wall directly from volumes of 2D CT slices and generates its segmentation masks from the predicted 3D point cloud. Extensive experimental results show that by incorporating a shape prior from the point cloud, the segmentation masks are more accurate than the state-of-the-art U-Net results in terms of Dice's coefficient and Hausdorff distance.The proposed joint learning framework of our PC-U net is beneficial for automatic cardiac image analysis tasks because it can obtain simultaneously the 3D shape and segmentation of the LV MYO walls.
Weakly Supervised Deep Nuclei Segmentation Using Partial Points Annotation in Histopathology Images
Jul 10, 2020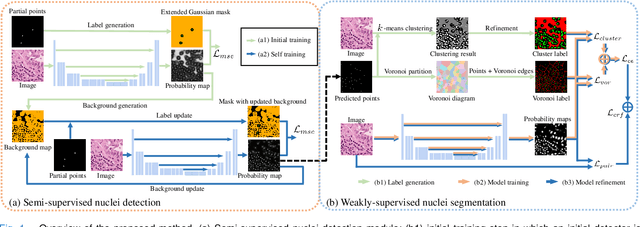
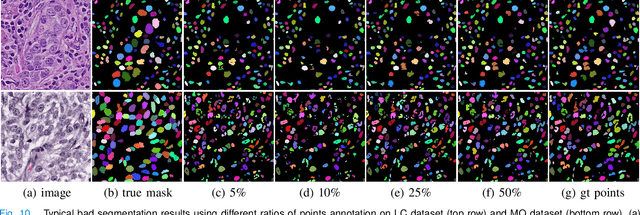
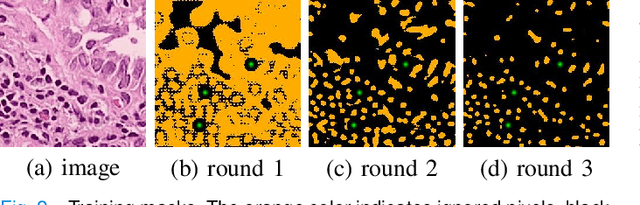
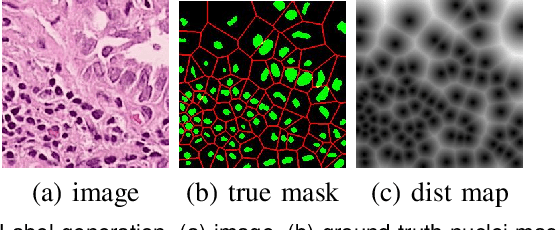
Abstract:Nuclei segmentation is a fundamental task in histopathology image analysis. Typically, such segmentation tasks require significant effort to manually generate accurate pixel-wise annotations for fully supervised training. To alleviate such tedious and manual effort, in this paper we propose a novel weakly supervised segmentation framework based on partial points annotation, i.e., only a small portion of nuclei locations in each image are labeled. The framework consists of two learning stages. In the first stage, we design a semi-supervised strategy to learn a detection model from partially labeled nuclei locations. Specifically, an extended Gaussian mask is designed to train an initial model with partially labeled data. Then, selftraining with background propagation is proposed to make use of the unlabeled regions to boost nuclei detection and suppress false positives. In the second stage, a segmentation model is trained from the detected nuclei locations in a weakly-supervised fashion. Two types of coarse labels with complementary information are derived from the detected points and are then utilized to train a deep neural network. The fully-connected conditional random field loss is utilized in training to further refine the model without introducing extra computational complexity during inference. The proposed method is extensively evaluated on two nuclei segmentation datasets. The experimental results demonstrate that our method can achieve competitive performance compared to the fully supervised counterpart and the state-of-the-art methods while requiring significantly less annotation effort.
Vertebra-Focused Landmark Detection for Scoliosis Assessment
Jan 09, 2020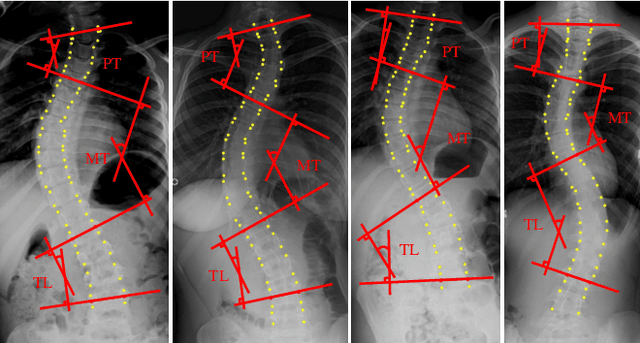

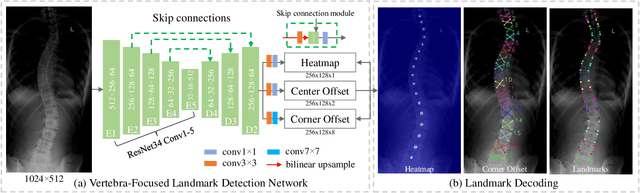
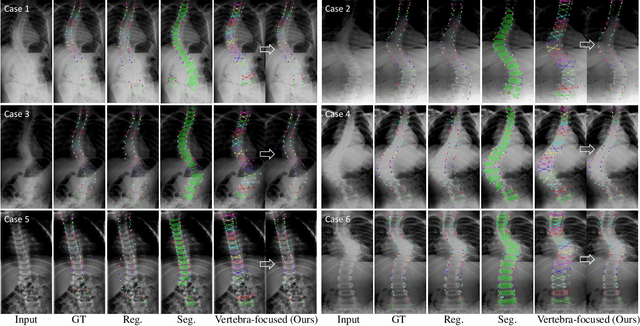
Abstract:Adolescent idiopathic scoliosis (AIS) is a lifetime disease that arises in children. Accurate estimation of Cobb angles of the scoliosis is essential for clinicians to make diagnosis and treatment decisions. The Cobb angles are measured according to the vertebrae landmarks. Existing regression-based methods for the vertebra landmark detection typically suffer from large dense mapping parameters and inaccurate landmark localization. The segmentation-based methods tend to predict connected or corrupted vertebra masks. In this paper, we propose a novel vertebra-focused landmark detection method. Our model first localizes the vertebra centers, based on which it then traces the four corner landmarks of the vertebra through the learned corner offset. In this way, our method is able to keep the order of the landmarks. The comparison results demonstrate the merits of our method in both Cobb angle measurement and landmark detection on low-contrast and ambiguous X-ray images. Code is available at: \url{https://github.com/yijingru/Vertebra-Landmark-Detection}.
Point Cloud Processing via Recurrent Set Encoding
Nov 25, 2019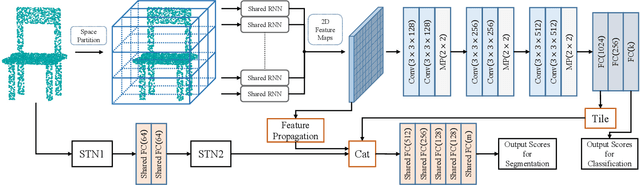
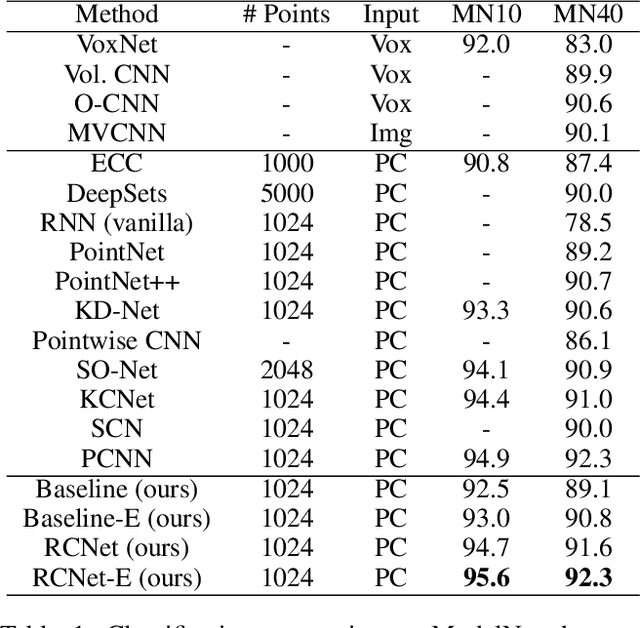
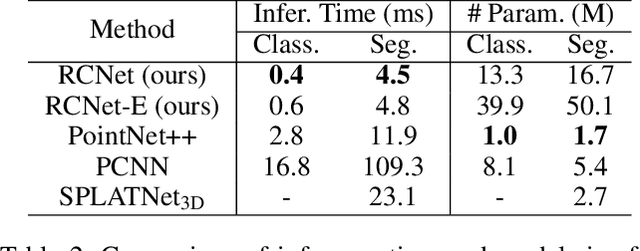
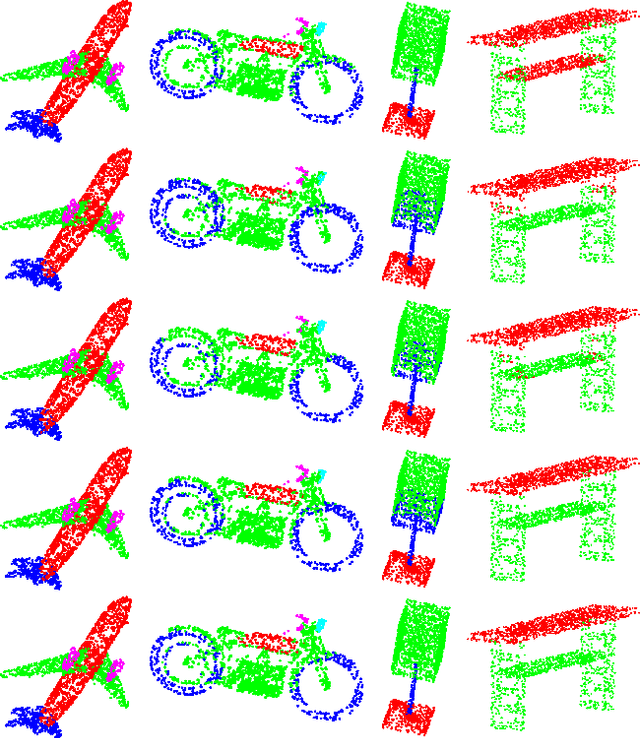
Abstract:We present a new permutation-invariant network for 3D point cloud processing. Our network is composed of a recurrent set encoder and a convolutional feature aggregator. Given an unordered point set, the encoder firstly partitions its ambient space into parallel beams. Points within each beam are then modeled as a sequence and encoded into subregional geometric features by a shared recurrent neural network (RNN). The spatial layout of the beams is regular, and this allows the beam features to be further fed into an efficient 2D convolutional neural network (CNN) for hierarchical feature aggregation. Our network is effective at spatial feature learning, and competes favorably with the state-of-the-arts (SOTAs) on a number of benchmarks. Meanwhile, it is significantly more efficient compared to the SOTAs.
Object-Guided Instance Segmentation for Biological Images
Nov 20, 2019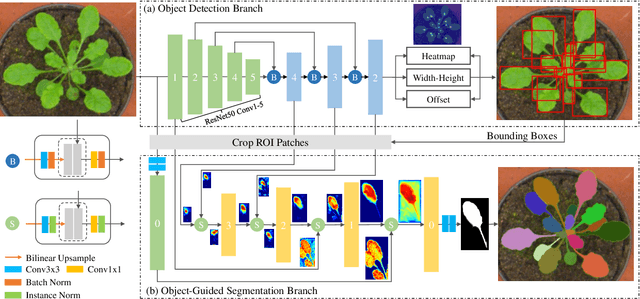
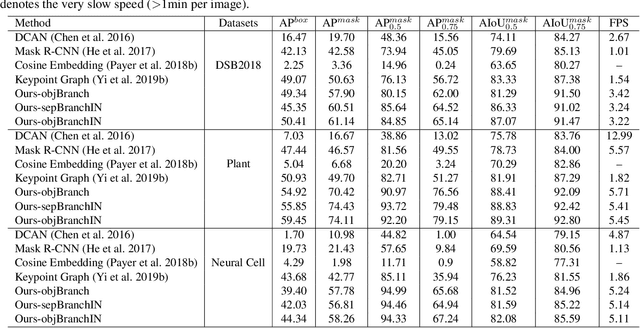
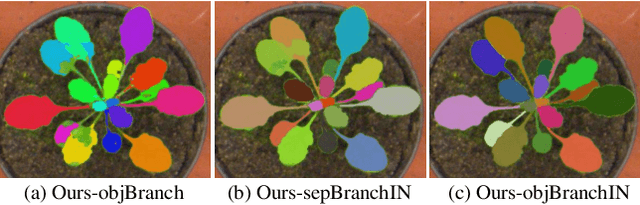
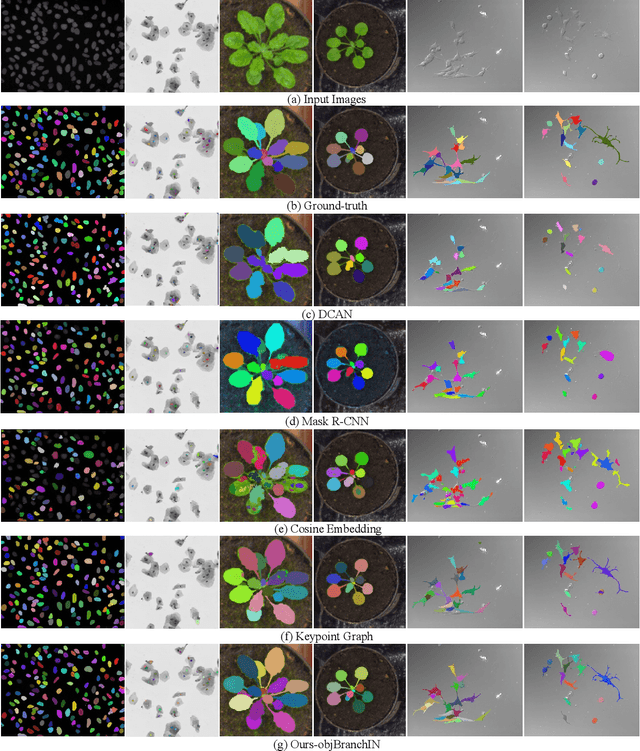
Abstract:Instance segmentation of biological images is essential for studying object behaviors and properties. The challenges, such as clustering, occlusion, and adhesion problems of the objects, make instance segmentation a non-trivial task. Current box-free instance segmentation methods typically rely on local pixel-level information. Due to a lack of global object view, these methods are prone to over- or under-segmentation. On the contrary, the box-based instance segmentation methods incorporate object detection into the segmentation, performing better in identifying the individual instances. In this paper, we propose a new box-based instance segmentation method. Mainly, we locate the object bounding boxes from their center points. The object features are subsequently reused in the segmentation branch as a guide to separate the clustered instances within an RoI patch. Along with the instance normalization, the model is able to recover the target object distribution and suppress the distribution of neighboring attached objects. Consequently, the proposed model performs excellently in segmenting the clustered objects while retaining the target object details. The proposed method achieves state-of-the-art performances on three biological datasets: cell nuclei, plant phenotyping dataset, and neural cells.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge