Ian Adelstein
HiPoNet: A Topology-Preserving Multi-View Neural Network For High Dimensional Point Cloud and Single-Cell Data
Feb 11, 2025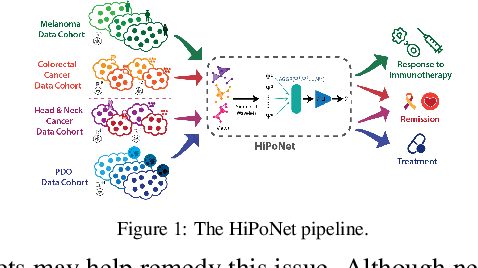
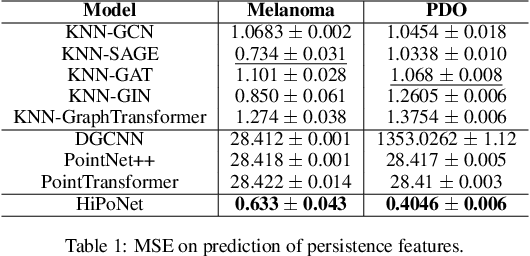
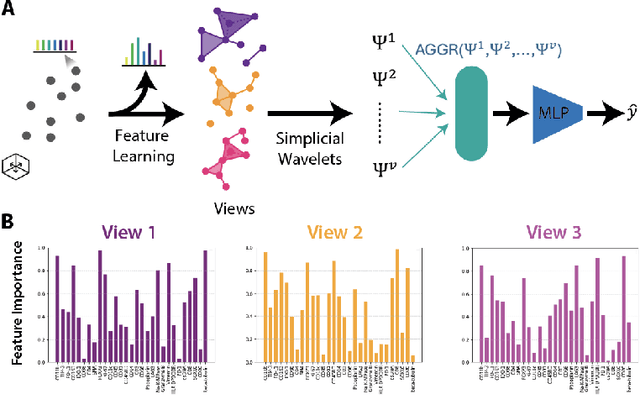
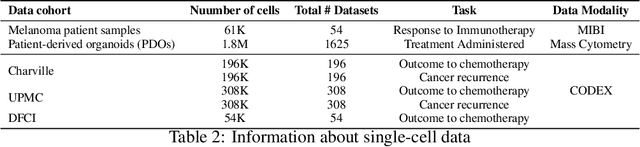
Abstract:In this paper, we propose HiPoNet, an end-to-end differentiable neural network for regression, classification, and representation learning on high-dimensional point clouds. Single-cell data can have high dimensionality exceeding the capabilities of existing methods point cloud tailored for 3D data. Moreover, modern single-cell and spatial experiments now yield entire cohorts of datasets (i.e. one on every patient), necessitating models that can process large, high-dimensional point clouds at scale. Most current approaches build a single nearest-neighbor graph, discarding important geometric information. In contrast, HiPoNet forms higher-order simplicial complexes through learnable feature reweighting, generating multiple data views that disentangle distinct biological processes. It then employs simplicial wavelet transforms to extract multi-scale features - capturing both local and global topology. We empirically show that these components preserve topological information in the learned representations, and that HiPoNet significantly outperforms state-of-the-art point-cloud and graph-based models on single cell. We also show an application of HiPoNet on spatial transcriptomics datasets using spatial co-ordinates as one of the views. Overall, HiPoNet offers a robust and scalable solution for high-dimensional data analysis.
Exploring the Manifold of Neural Networks Using Diffusion Geometry
Nov 19, 2024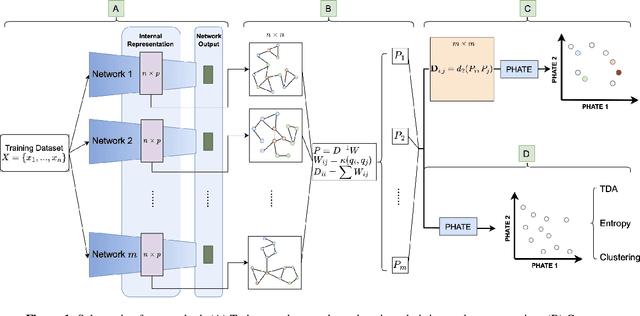

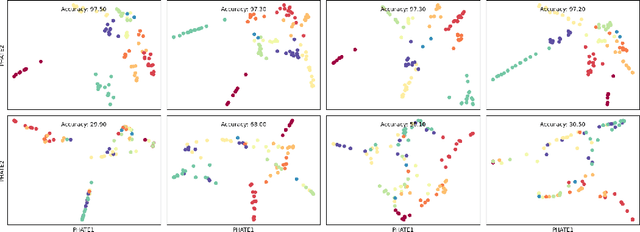
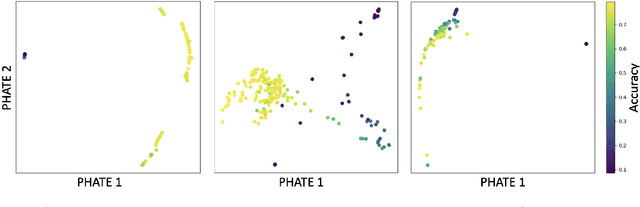
Abstract:Drawing motivation from the manifold hypothesis, which posits that most high-dimensional data lies on or near low-dimensional manifolds, we apply manifold learning to the space of neural networks. We learn manifolds where datapoints are neural networks by introducing a distance between the hidden layer representations of the neural networks. These distances are then fed to the non-linear dimensionality reduction algorithm PHATE to create a manifold of neural networks. We characterize this manifold using features of the representation, including class separation, hierarchical cluster structure, spectral entropy, and topological structure. Our analysis reveals that high-performing networks cluster together in the manifold, displaying consistent embedding patterns across all these features. Finally, we demonstrate the utility of this approach for guiding hyperparameter optimization and neural architecture search by sampling from the manifold.
Geometry-Aware Generative Autoencoders for Warped Riemannian Metric Learning and Generative Modeling on Data Manifolds
Oct 16, 2024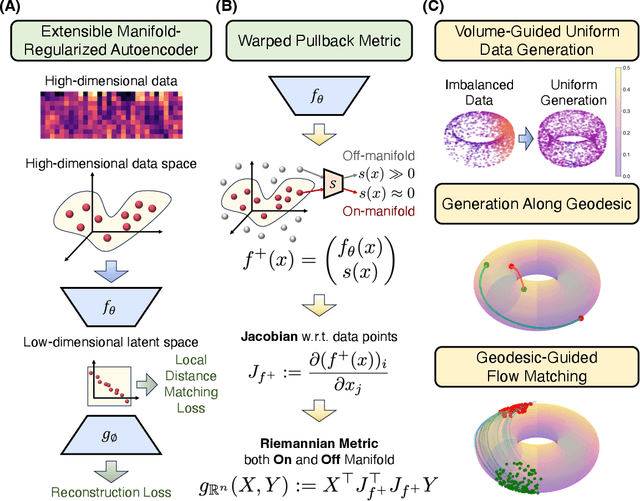

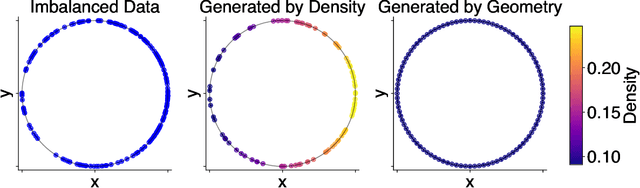
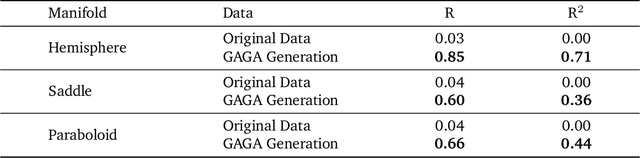
Abstract:Rapid growth of high-dimensional datasets in fields such as single-cell RNA sequencing and spatial genomics has led to unprecedented opportunities for scientific discovery, but it also presents unique computational and statistical challenges. Traditional methods struggle with geometry-aware data generation, interpolation along meaningful trajectories, and transporting populations via feasible paths. To address these issues, we introduce Geometry-Aware Generative Autoencoder (GAGA), a novel framework that combines extensible manifold learning with generative modeling. GAGA constructs a neural network embedding space that respects the intrinsic geometries discovered by manifold learning and learns a novel warped Riemannian metric on the data space. This warped metric is derived from both the points on the data manifold and negative samples off the manifold, allowing it to characterize a meaningful geometry across the entire latent space. Using this metric, GAGA can uniformly sample points on the manifold, generate points along geodesics, and interpolate between populations across the learned manifold. GAGA shows competitive performance in simulated and real world datasets, including a 30% improvement over the state-of-the-art methods in single-cell population-level trajectory inference.
Assessing Neural Network Representations During Training Using Noise-Resilient Diffusion Spectral Entropy
Dec 04, 2023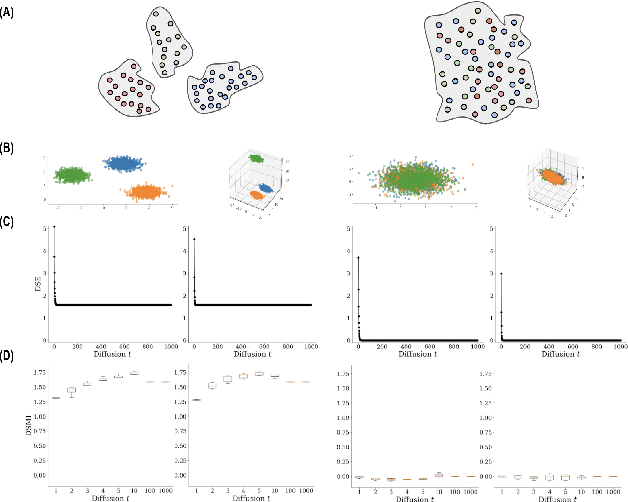

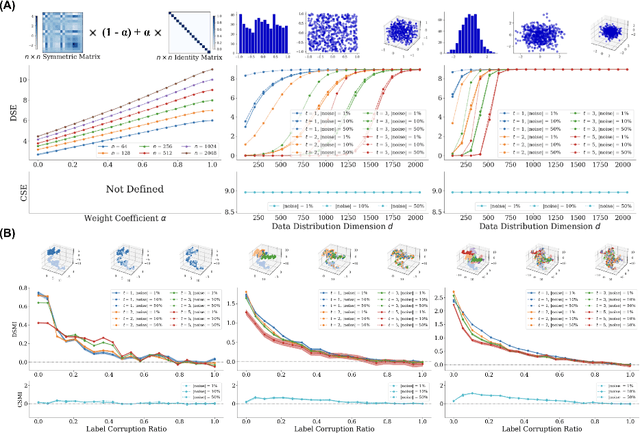

Abstract:Entropy and mutual information in neural networks provide rich information on the learning process, but they have proven difficult to compute reliably in high dimensions. Indeed, in noisy and high-dimensional data, traditional estimates in ambient dimensions approach a fixed entropy and are prohibitively hard to compute. To address these issues, we leverage data geometry to access the underlying manifold and reliably compute these information-theoretic measures. Specifically, we define diffusion spectral entropy (DSE) in neural representations of a dataset as well as diffusion spectral mutual information (DSMI) between different variables representing data. First, we show that they form noise-resistant measures of intrinsic dimensionality and relationship strength in high-dimensional simulated data that outperform classic Shannon entropy, nonparametric estimation, and mutual information neural estimation (MINE). We then study the evolution of representations in classification networks with supervised learning, self-supervision, or overfitting. We observe that (1) DSE of neural representations increases during training; (2) DSMI with the class label increases during generalizable learning but stays stagnant during overfitting; (3) DSMI with the input signal shows differing trends: on MNIST it increases, while on CIFAR-10 and STL-10 it decreases. Finally, we show that DSE can be used to guide better network initialization and that DSMI can be used to predict downstream classification accuracy across 962 models on ImageNet. The official implementation is available at https://github.com/ChenLiu-1996/DiffusionSpectralEntropy.
BLIS-Net: Classifying and Analyzing Signals on Graphs
Oct 26, 2023Abstract:Graph neural networks (GNNs) have emerged as a powerful tool for tasks such as node classification and graph classification. However, much less work has been done on signal classification, where the data consists of many functions (referred to as signals) defined on the vertices of a single graph. These tasks require networks designed differently from those designed for traditional GNN tasks. Indeed, traditional GNNs rely on localized low-pass filters, and signals of interest may have intricate multi-frequency behavior and exhibit long range interactions. This motivates us to introduce the BLIS-Net (Bi-Lipschitz Scattering Net), a novel GNN that builds on the previously introduced geometric scattering transform. Our network is able to capture both local and global signal structure and is able to capture both low-frequency and high-frequency information. We make several crucial changes to the original geometric scattering architecture which we prove increase the ability of our network to capture information about the input signal and show that BLIS-Net achieves superior performance on both synthetic and real-world data sets based on traffic flow and fMRI data.
A Flow Artist for High-Dimensional Cellular Data
Jul 31, 2023Abstract:We consider the problem of embedding point cloud data sampled from an underlying manifold with an associated flow or velocity. Such data arises in many contexts where static snapshots of dynamic entities are measured, including in high-throughput biology such as single-cell transcriptomics. Existing embedding techniques either do not utilize velocity information or embed the coordinates and velocities independently, i.e., they either impose velocities on top of an existing point embedding or embed points within a prescribed vector field. Here we present FlowArtist, a neural network that embeds points while jointly learning a vector field around the points. The combination allows FlowArtist to better separate and visualize velocity-informed structures. Our results, on toy datasets and single-cell RNA velocity data, illustrate the value of utilizing coordinate and velocity information in tandem for embedding and visualizing high-dimensional data.
Neural FIM for learning Fisher Information Metrics from point cloud data
Jun 12, 2023Abstract:Although data diffusion embeddings are ubiquitous in unsupervised learning and have proven to be a viable technique for uncovering the underlying intrinsic geometry of data, diffusion embeddings are inherently limited due to their discrete nature. To this end, we propose neural FIM, a method for computing the Fisher information metric (FIM) from point cloud data - allowing for a continuous manifold model for the data. Neural FIM creates an extensible metric space from discrete point cloud data such that information from the metric can inform us of manifold characteristics such as volume and geodesics. We demonstrate Neural FIM's utility in selecting parameters for the PHATE visualization method as well as its ability to obtain information pertaining to local volume illuminating branching points and cluster centers embeddings of a toy dataset and two single-cell datasets of IPSC reprogramming and PBMCs (immune cells).
A Heat Diffusion Perspective on Geodesic Preserving Dimensionality Reduction
May 30, 2023Abstract:Diffusion-based manifold learning methods have proven useful in representation learning and dimensionality reduction of modern high dimensional, high throughput, noisy datasets. Such datasets are especially present in fields like biology and physics. While it is thought that these methods preserve underlying manifold structure of data by learning a proxy for geodesic distances, no specific theoretical links have been established. Here, we establish such a link via results in Riemannian geometry explicitly connecting heat diffusion to manifold distances. In this process, we also formulate a more general heat kernel based manifold embedding method that we call heat geodesic embeddings. This novel perspective makes clearer the choices available in manifold learning and denoising. Results show that our method outperforms existing state of the art in preserving ground truth manifold distances, and preserving cluster structure in toy datasets. We also showcase our method on single cell RNA-sequencing datasets with both continuum and cluster structure, where our method enables interpolation of withheld timepoints of data. Finally, we show that parameters of our more general method can be configured to give results similar to PHATE (a state-of-the-art diffusion based manifold learning method) as well as SNE (an attraction/repulsion neighborhood based method that forms the basis of t-SNE).
Diffusion Curvature for Estimating Local Curvature in High Dimensional Data
Jun 08, 2022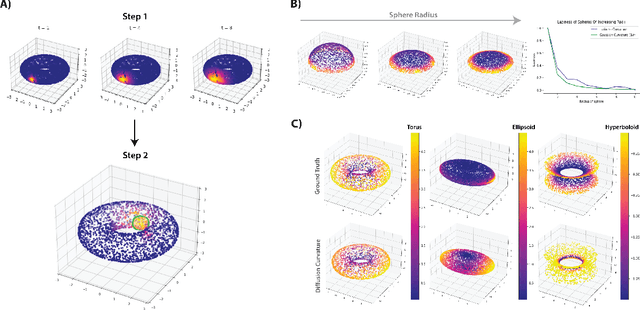
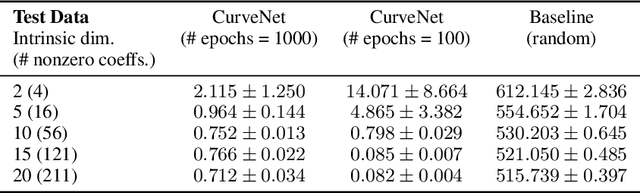
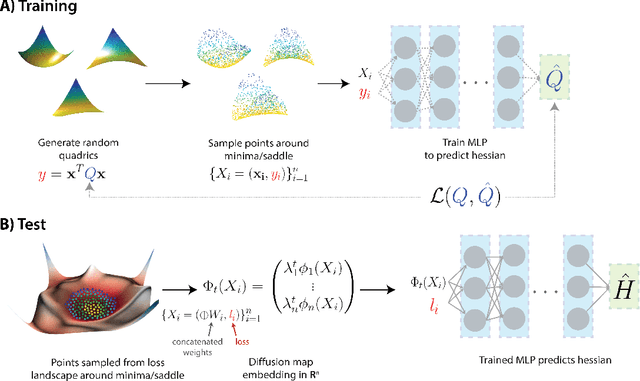
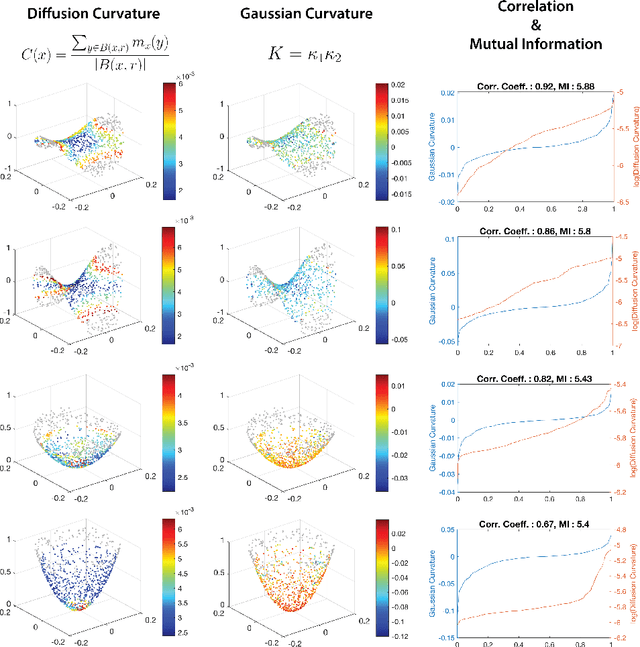
Abstract:We introduce a new intrinsic measure of local curvature on point-cloud data called diffusion curvature. Our measure uses the framework of diffusion maps, including the data diffusion operator, to structure point cloud data and define local curvature based on the laziness of a random walk starting at a point or region of the data. We show that this laziness directly relates to volume comparison results from Riemannian geometry. We then extend this scalar curvature notion to an entire quadratic form using neural network estimations based on the diffusion map of point-cloud data. We show applications of both estimations on toy data, single-cell data, and on estimating local Hessian matrices of neural network loss landscapes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge