Hyunseok Lee
Vision-aligned Latent Reasoning for Multi-modal Large Language Model
Feb 04, 2026Abstract:Despite recent advancements in Multi-modal Large Language Models (MLLMs) on diverse understanding tasks, these models struggle to solve problems which require extensive multi-step reasoning. This is primarily due to the progressive dilution of visual information during long-context generation, which hinders their ability to fully exploit test-time scaling. To address this issue, we introduce Vision-aligned Latent Reasoning (VaLR), a simple, yet effective reasoning framework that dynamically generates vision-aligned latent tokens before each Chain of Thought reasoning step, guiding the model to reason based on perceptual cues in the latent space. Specifically, VaLR is trained to preserve visual knowledge during reasoning by aligning intermediate embeddings of MLLM with those from vision encoders. Empirical results demonstrate that VaLR consistently outperforms existing approaches across a wide range of benchmarks requiring long-context understanding or precise visual perception, while exhibiting test-time scaling behavior not observed in prior MLLMs. In particular, VaLR improves the performance significantly from 33.0% to 52.9% on VSI-Bench, achieving a 19.9%p gain over Qwen2.5-VL.
ReGUIDE: Data Efficient GUI Grounding via Spatial Reasoning and Search
May 21, 2025Abstract:Recent advances in Multimodal Large Language Models (MLLMs) have enabled autonomous agents to interact with computers via Graphical User Interfaces (GUIs), where accurately localizing the coordinates of interface elements (e.g., buttons) is often required for fine-grained actions. However, this remains significantly challenging, leading prior works to rely on large-scale web datasets to improve the grounding accuracy. In this work, we propose Reasoning Graphical User Interface Grounding for Data Efficiency (ReGUIDE), a novel and effective framework for web grounding that enables MLLMs to learn data efficiently through self-generated reasoning and spatial-aware criticism. More specifically, ReGUIDE learns to (i) self-generate a language reasoning process for the localization via online reinforcement learning, and (ii) criticize the prediction using spatial priors that enforce equivariance under input transformations. At inference time, ReGUIDE further boosts performance through a test-time scaling strategy, which combines spatial search with coordinate aggregation. Our experiments demonstrate that ReGUIDE significantly advances web grounding performance across multiple benchmarks, outperforming baselines with substantially fewer training data points (e.g., only 0.2% samples compared to the best open-sourced baselines).
ReVISE: Learning to Refine at Test-Time via Intrinsic Self-Verification
Feb 20, 2025Abstract:Self-awareness, i.e., the ability to assess and correct one's own generation, is a fundamental aspect of human intelligence, making its replication in large language models (LLMs) an important yet challenging task. Previous works tackle this by employing extensive reinforcement learning or rather relying on large external verifiers. In this work, we propose Refine via Intrinsic Self-Verification (ReVISE), an efficient and effective framework that enables LLMs to self-correct their outputs through self-verification. The core idea of ReVISE is to enable LLMs to verify their reasoning processes and continually rethink reasoning trajectories based on its verification. We introduce a structured curriculum based upon online preference learning to implement this efficiently. Specifically, as ReVISE involves two challenging tasks (i.e., self-verification and reasoning correction), we tackle each task sequentially using curriculum learning, collecting both failed and successful reasoning paths to construct preference pairs for efficient training. During inference, our approach enjoys natural test-time scaling by integrating self-verification and correction capabilities, further enhanced by our proposed confidence-aware decoding mechanism. Our experiments on various reasoning tasks demonstrate that ReVISE achieves efficient self-correction and significantly improves reasoning performance.
Deep Learning Techniques for Automatic Lateral X-ray Cephalometric Landmark Detection: Is the Problem Solved?
Sep 24, 2024Abstract:Localization of the craniofacial landmarks from lateral cephalograms is a fundamental task in cephalometric analysis. The automation of the corresponding tasks has thus been the subject of intense research over the past decades. In this paper, we introduce the "Cephalometric Landmark Detection (CL-Detection)" dataset, which is the largest publicly available and comprehensive dataset for cephalometric landmark detection. This multi-center and multi-vendor dataset includes 600 lateral X-ray images with 38 landmarks acquired with different equipment from three medical centers. The overarching objective of this paper is to measure how far state-of-the-art deep learning methods can go for cephalometric landmark detection. Following the 2023 MICCAI CL-Detection Challenge, we report the results of the top ten research groups using deep learning methods. Results show that the best methods closely approximate the expert analysis, achieving a mean detection rate of 75.719% and a mean radial error of 1.518 mm. While there is room for improvement, these findings undeniably open the door to highly accurate and fully automatic location of craniofacial landmarks. We also identify scenarios for which deep learning methods are still failing. Both the dataset and detailed results are publicly available online, while the platform will remain open for the community to benchmark future algorithm developments at https://cl-detection2023.grand-challenge.org/.
ReMoDetect: Reward Models Recognize Aligned LLM's Generations
May 27, 2024Abstract:The remarkable capabilities and easy accessibility of large language models (LLMs) have significantly increased societal risks (e.g., fake news generation), necessitating the development of LLM-generated text (LGT) detection methods for safe usage. However, detecting LGTs is challenging due to the vast number of LLMs, making it impractical to account for each LLM individually; hence, it is crucial to identify the common characteristics shared by these models. In this paper, we draw attention to a common feature of recent powerful LLMs, namely the alignment training, i.e., training LLMs to generate human-preferable texts. Our key finding is that as these aligned LLMs are trained to maximize the human preferences, they generate texts with higher estimated preferences even than human-written texts; thus, such texts are easily detected by using the reward model (i.e., an LLM trained to model human preference distribution). Based on this finding, we propose two training schemes to further improve the detection ability of the reward model, namely (i) continual preference fine-tuning to make the reward model prefer aligned LGTs even further and (ii) reward modeling of Human/LLM mixed texts (a rephrased texts from human-written texts using aligned LLMs), which serves as a median preference text corpus between LGTs and human-written texts to learn the decision boundary better. We provide an extensive evaluation by considering six text domains across twelve aligned LLMs, where our method demonstrates state-of-the-art results. Code is available at https://github.com/hyunseoklee-ai/reward_llm_detect.
Assessing generalisability of deep learning-based polyp detection and segmentation methods through a computer vision challenge
Feb 24, 2022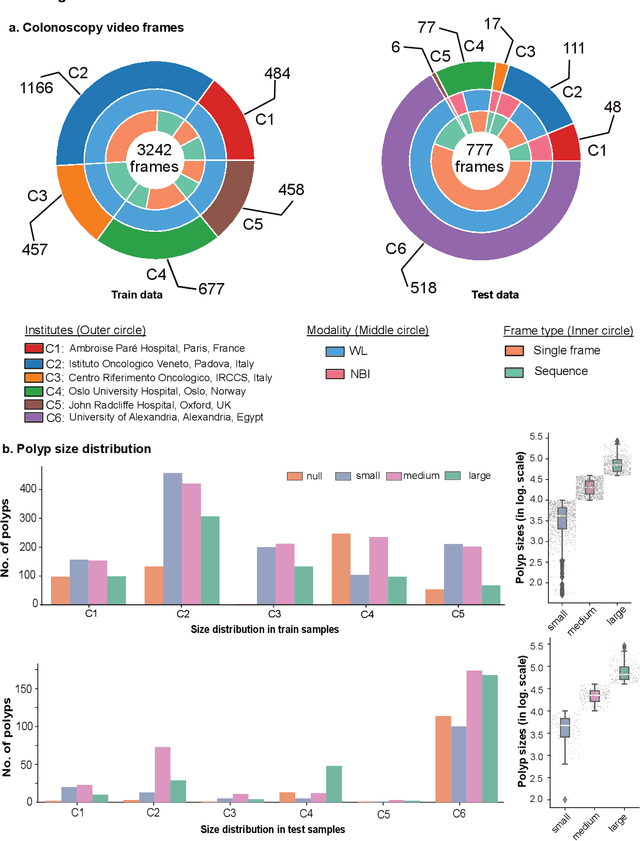
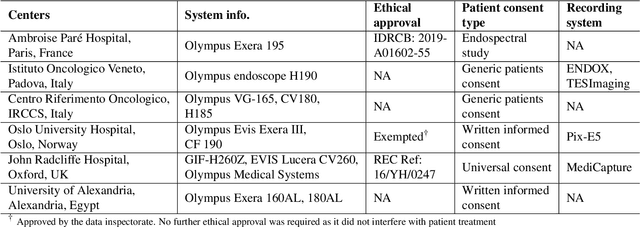
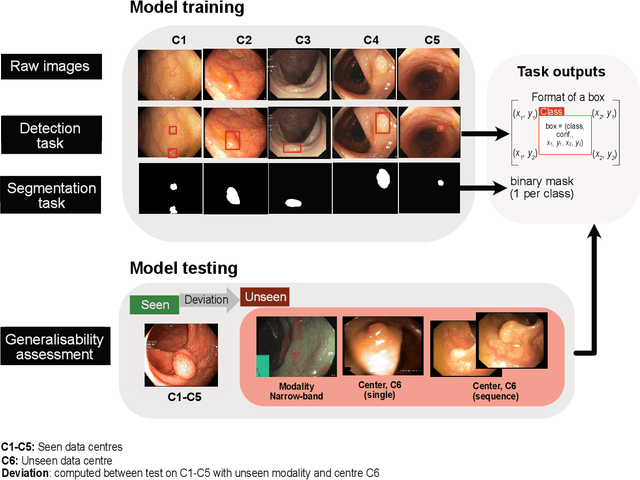
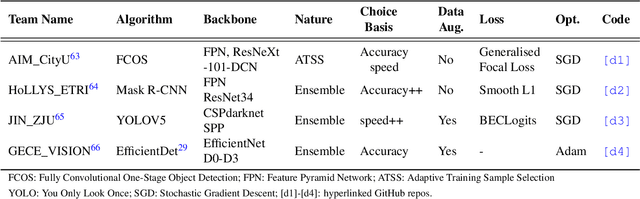
Abstract:Polyps are well-known cancer precursors identified by colonoscopy. However, variability in their size, location, and surface largely affect identification, localisation, and characterisation. Moreover, colonoscopic surveillance and removal of polyps (referred to as polypectomy ) are highly operator-dependent procedures. There exist a high missed detection rate and incomplete removal of colonic polyps due to their variable nature, the difficulties to delineate the abnormality, the high recurrence rates, and the anatomical topography of the colon. There have been several developments in realising automated methods for both detection and segmentation of these polyps using machine learning. However, the major drawback in most of these methods is their ability to generalise to out-of-sample unseen datasets that come from different centres, modalities and acquisition systems. To test this hypothesis rigorously we curated a multi-centre and multi-population dataset acquired from multiple colonoscopy systems and challenged teams comprising machine learning experts to develop robust automated detection and segmentation methods as part of our crowd-sourcing Endoscopic computer vision challenge (EndoCV) 2021. In this paper, we analyse the detection results of the four top (among seven) teams and the segmentation results of the five top teams (among 16). Our analyses demonstrate that the top-ranking teams concentrated on accuracy (i.e., accuracy > 80% on overall Dice score on different validation sets) over real-time performance required for clinical applicability. We further dissect the methods and provide an experiment-based hypothesis that reveals the need for improved generalisability to tackle diversity present in multi-centre datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge