Hyojin Kim
Generalizing AI-driven Assessment of Immunohistochemistry across Immunostains and Cancer Types: A Universal Immunohistochemistry Analyzer
Jul 30, 2024Abstract:Despite advancements in methodologies, immunohistochemistry (IHC) remains the most utilized ancillary test for histopathologic and companion diagnostics in targeted therapies. However, objective IHC assessment poses challenges. Artificial intelligence (AI) has emerged as a potential solution, yet its development requires extensive training for each cancer and IHC type, limiting versatility. We developed a Universal IHC (UIHC) analyzer, an AI model for interpreting IHC images regardless of tumor or IHC types, using training datasets from various cancers stained for PD-L1 and/or HER2. This multi-cohort trained model outperforms conventional single-cohort models in interpreting unseen IHCs (Kappa score 0.578 vs. up to 0.509) and consistently shows superior performance across different positive staining cutoff values. Qualitative analysis reveals that UIHC effectively clusters patches based on expression levels. The UIHC model also quantitatively assesses c-MET expression with MET mutations, representing a significant advancement in AI application in the era of personalized medicine and accumulating novel biomarkers.
Advancing Cross-Domain Generalizability in Face Anti-Spoofing: Insights, Design, and Metrics
Jun 18, 2024



Abstract:This paper presents a novel perspective for enhancing anti-spoofing performance in zero-shot data domain generalization. Unlike traditional image classification tasks, face anti-spoofing datasets display unique generalization characteristics, necessitating novel zero-shot data domain generalization. One step forward to the previous frame-wise spoofing prediction, we introduce a nuanced metric calculation that aggregates frame-level probabilities for a video-wise prediction, to tackle the gap between the reported frame-wise accuracy and instability in real-world use-case. This approach enables the quantification of bias and variance in model predictions, offering a more refined analysis of model generalization. Our investigation reveals that simply scaling up the backbone of models does not inherently improve the mentioned instability, leading us to propose an ensembled backbone method from a Bayesian perspective. The probabilistically ensembled backbone both improves model robustness measured from the proposed metric and spoofing accuracy, and also leverages the advantages of measuring uncertainty, allowing for enhanced sampling during training that contributes to model generalization across new datasets. We evaluate the proposed method from the benchmark OMIC dataset and also the public CelebA-Spoof and SiW-Mv2. Our final model outperforms existing state-of-the-art methods across the datasets, showcasing advancements in Bias, Variance, HTER, and AUC metrics.
Distributed Stochastic Optimization of a Neural Representation Network for Time-Space Tomography Reconstruction
Apr 29, 2024



Abstract:4D time-space reconstruction of dynamic events or deforming objects using X-ray computed tomography (CT) is an extremely ill-posed inverse problem. Existing approaches assume that the object remains static for the duration of several tens or hundreds of X-ray projection measurement images (reconstruction of consecutive limited-angle CT scans). However, this is an unrealistic assumption for many in-situ experiments that causes spurious artifacts and inaccurate morphological reconstructions of the object. To solve this problem, we propose to perform a 4D time-space reconstruction using a distributed implicit neural representation (DINR) network that is trained using a novel distributed stochastic training algorithm. Our DINR network learns to reconstruct the object at its output by iterative optimization of its network parameters such that the measured projection images best match the output of the CT forward measurement model. We use a continuous time and space forward measurement model that is a function of the DINR outputs at a sparsely sampled set of continuous valued object coordinates. Unlike existing state-of-the-art neural representation architectures that forward and back propagate through dense voxel grids that sample the object's entire time-space coordinates, we only propagate through the DINR at a small subset of object coordinates in each iteration resulting in an order-of-magnitude reduction in memory and compute for training. DINR leverages distributed computation across several compute nodes and GPUs to produce high-fidelity 4D time-space reconstructions even for extremely large CT data sizes. We use both simulated parallel-beam and experimental cone-beam X-ray CT datasets to demonstrate the superior performance of our approach.
HyperCLOVA X Technical Report
Apr 13, 2024Abstract:We introduce HyperCLOVA X, a family of large language models (LLMs) tailored to the Korean language and culture, along with competitive capabilities in English, math, and coding. HyperCLOVA X was trained on a balanced mix of Korean, English, and code data, followed by instruction-tuning with high-quality human-annotated datasets while abiding by strict safety guidelines reflecting our commitment to responsible AI. The model is evaluated across various benchmarks, including comprehensive reasoning, knowledge, commonsense, factuality, coding, math, chatting, instruction-following, and harmlessness, in both Korean and English. HyperCLOVA X exhibits strong reasoning capabilities in Korean backed by a deep understanding of the language and cultural nuances. Further analysis of the inherent bilingual nature and its extension to multilingualism highlights the model's cross-lingual proficiency and strong generalization ability to untargeted languages, including machine translation between several language pairs and cross-lingual inference tasks. We believe that HyperCLOVA X can provide helpful guidance for regions or countries in developing their sovereign LLMs.
Differentiable Forward Projector for X-ray Computed Tomography
Jul 11, 2023



Abstract:Data-driven deep learning has been successfully applied to various computed tomographic reconstruction problems. The deep inference models may outperform existing analytical and iterative algorithms, especially in ill-posed CT reconstruction. However, those methods often predict images that do not agree with the measured projection data. This paper presents an accurate differentiable forward and back projection software library to ensure the consistency between the predicted images and the original measurements. The software library efficiently supports various projection geometry types while minimizing the GPU memory footprint requirement, which facilitates seamless integration with existing deep learning training and inference pipelines. The proposed software is available as open source: https://github.com/LLNL/LEAP.
DOLCE: A Model-Based Probabilistic Diffusion Framework for Limited-Angle CT Reconstruction
Nov 22, 2022Abstract:Limited-Angle Computed Tomography (LACT) is a non-destructive evaluation technique used in a variety of applications ranging from security to medicine. The limited angle coverage in LACT is often a dominant source of severe artifacts in the reconstructed images, making it a challenging inverse problem. We present DOLCE, a new deep model-based framework for LACT that uses a conditional diffusion model as an image prior. Diffusion models are a recent class of deep generative models that are relatively easy to train due to their implementation as image denoisers. DOLCE can form high-quality images from severely under-sampled data by integrating data-consistency updates with the sampling updates of a diffusion model, which is conditioned on the transformed limited-angle data. We show through extensive experimentation on several challenging real LACT datasets that, the same pre-trained DOLCE model achieves the SOTA performance on drastically different types of images. Additionally, we show that, unlike standard LACT reconstruction methods, DOLCE naturally enables the quantification of the reconstruction uncertainty by generating multiple samples consistent with the measured data.
Dynamic CT Reconstruction from Limited Views with Implicit Neural Representations and Parametric Motion Fields
Apr 23, 2021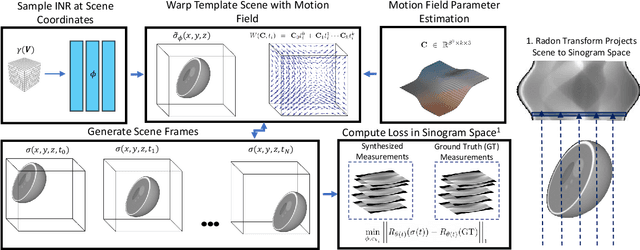
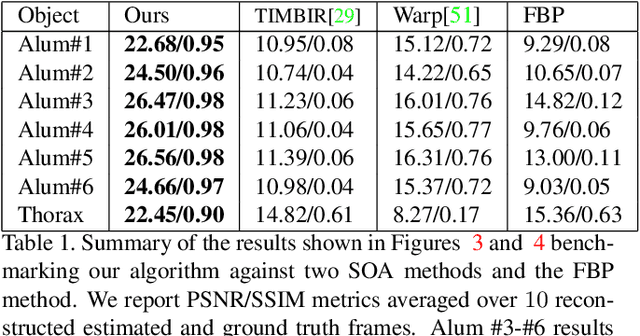
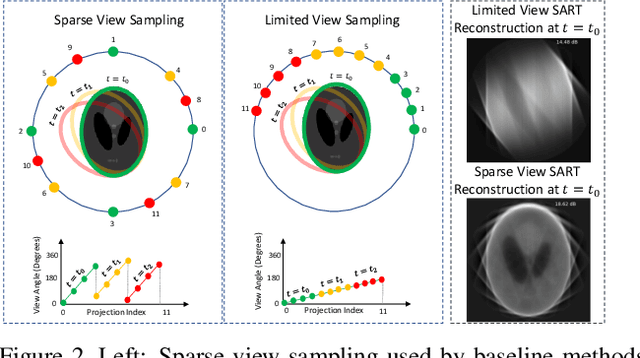
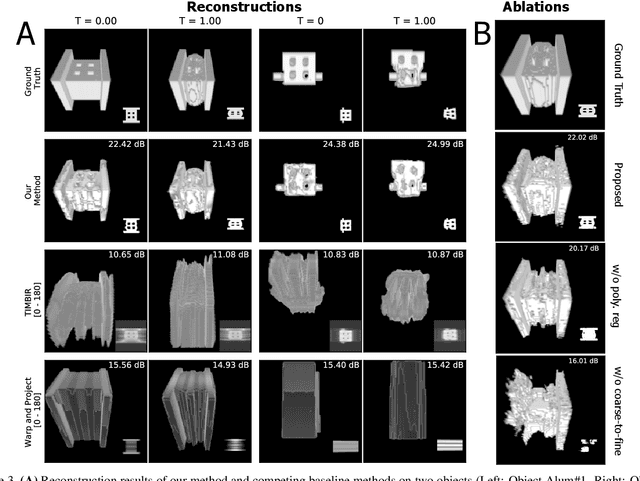
Abstract:Reconstructing dynamic, time-varying scenes with computed tomography (4D-CT) is a challenging and ill-posed problem common to industrial and medical settings. Existing 4D-CT reconstructions are designed for sparse sampling schemes that require fast CT scanners to capture multiple, rapid revolutions around the scene in order to generate high quality results. However, if the scene is moving too fast, then the sampling occurs along a limited view and is difficult to reconstruct due to spatiotemporal ambiguities. In this work, we design a reconstruction pipeline using implicit neural representations coupled with a novel parametric motion field warping to perform limited view 4D-CT reconstruction of rapidly deforming scenes. Importantly, we utilize a differentiable analysis-by-synthesis approach to compare with captured x-ray sinogram data in a self-supervised fashion. Thus, our resulting optimization method requires no training data to reconstruct the scene. We demonstrate that our proposed system robustly reconstructs scenes containing deformable and periodic motion and validate against state-of-the-art baselines. Further, we demonstrate an ability to reconstruct continuous spatiotemporal representations of our scenes and upsample them to arbitrary volumes and frame rates post-optimization. This research opens a new avenue for implicit neural representations in computed tomography reconstruction in general.
High-Throughput Virtual Screening of Small Molecule Inhibitors for SARS-CoV-2 Protein Targets with Deep Fusion Models
Apr 09, 2021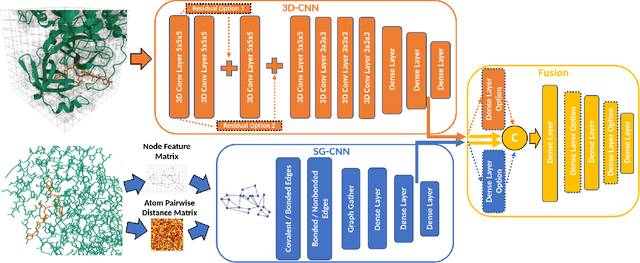
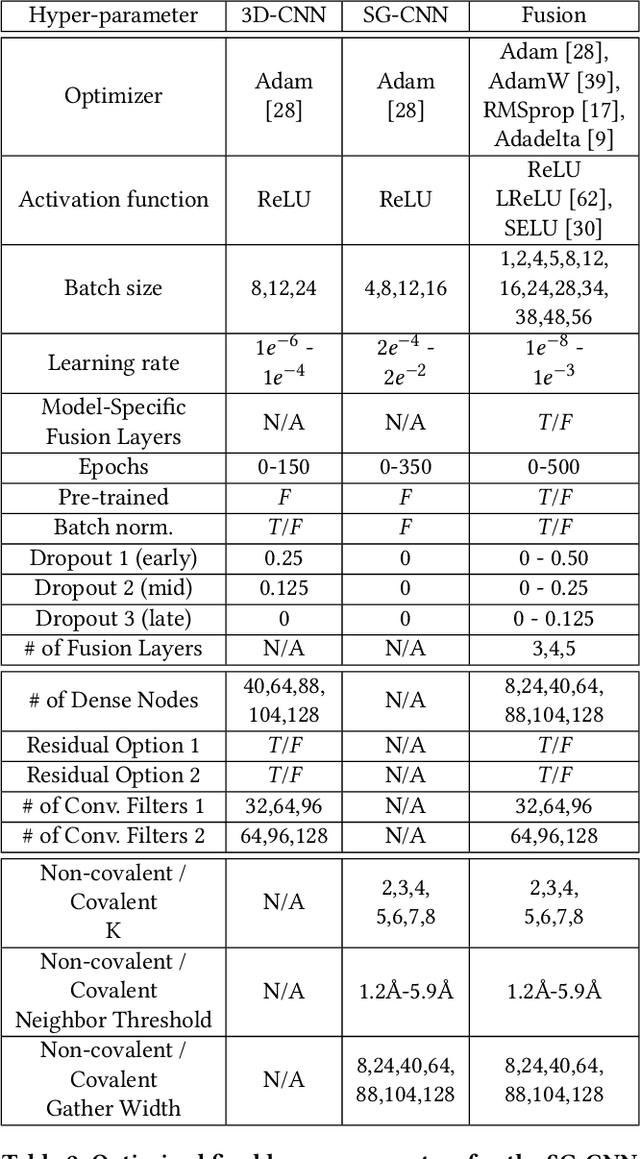
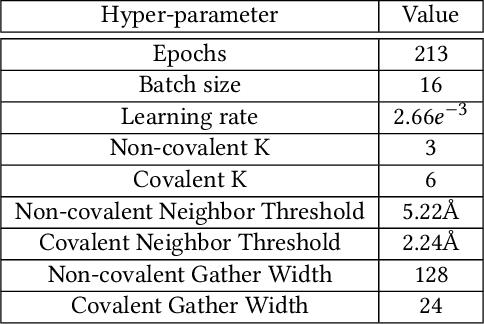
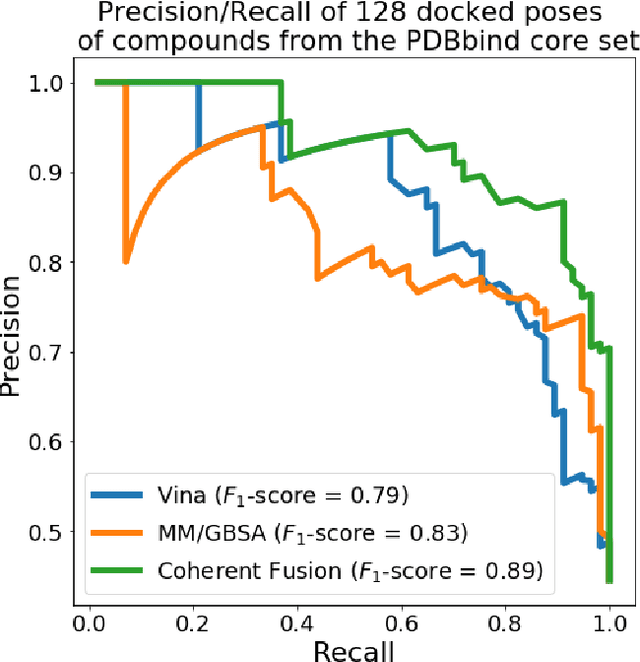
Abstract:Structure-based Deep Fusion models were recently shown to outperform several physics- and machine learning-based protein-ligand binding affinity prediction methods. As part of a multi-institutional COVID-19 pandemic response, over 500 million small molecules were computationally screened against four protein structures from the novel coronavirus (SARS-CoV-2), which causes COVID-19. Three enhancements to Deep Fusion were made in order to evaluate more than 5 billion docked poses on SARS-CoV-2 protein targets. First, the Deep Fusion concept was refined by formulating the architecture as one, coherently backpropagated model (Coherent Fusion) to improve binding-affinity prediction accuracy. Secondly, the model was trained using a distributed, genetic hyper-parameter optimization. Finally, a scalable, high-throughput screening capability was developed to maximize the number of ligands evaluated and expedite the path to experimental evaluation. In this work, we present both the methods developed for machine learning-based high-throughput screening and results from using our computational pipeline to find SARS-CoV-2 inhibitors.
Improved Protein-ligand Binding Affinity Prediction with Structure-Based Deep Fusion Inference
May 17, 2020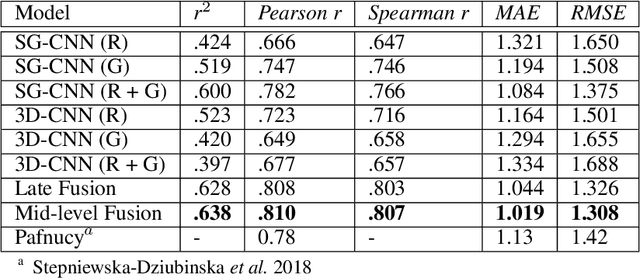
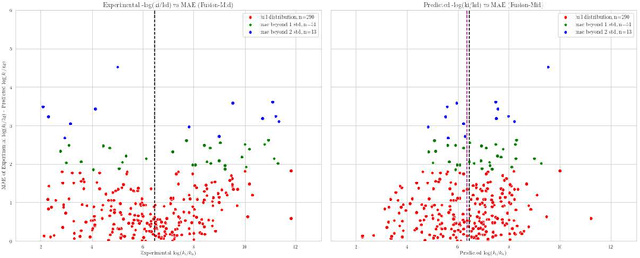
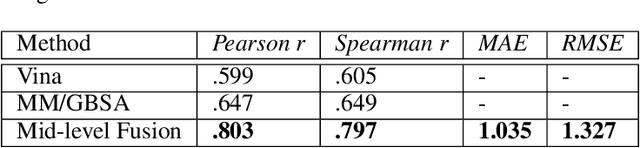
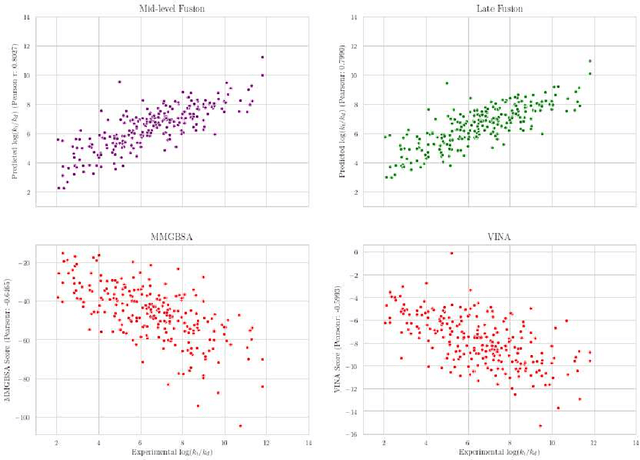
Abstract:Predicting accurate protein-ligand binding affinity is important in drug discovery but remains a challenge even with computationally expensive biophysics-based energy scoring methods and state-of-the-art deep learning approaches. Despite the recent advances in the deep convolutional and graph neural network based approaches, the model performance depends on the input data representation and suffers from distinct limitations. It is natural to combine complementary features and their inference from the individual models for better predictions. We present fusion models to benefit from different feature representations of two neural network models to improve the binding affinity prediction. We demonstrate effectiveness of the proposed approach by performing experiments with the PDBBind 2016 dataset and its docking pose complexes. The results show that the proposed approach improves the overall prediction compared to the individual neural network models with greater computational efficiency than related biophysics based energy scoring functions. We also discuss the benefit of the proposed fusion inference with several example complexes. The software is made available as open source at https://github.com/llnl/fast.
Improving Limited Angle CT Reconstruction with a Robust GAN Prior
Oct 14, 2019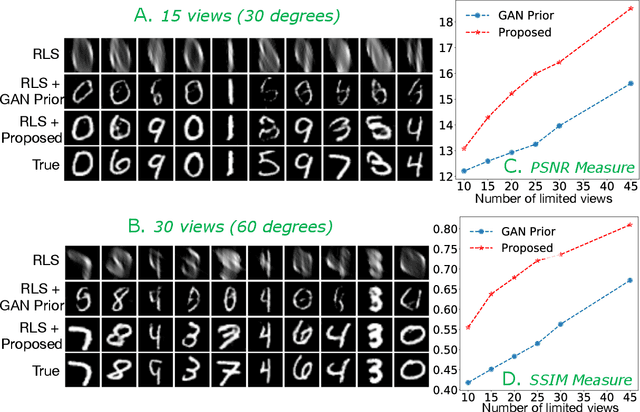
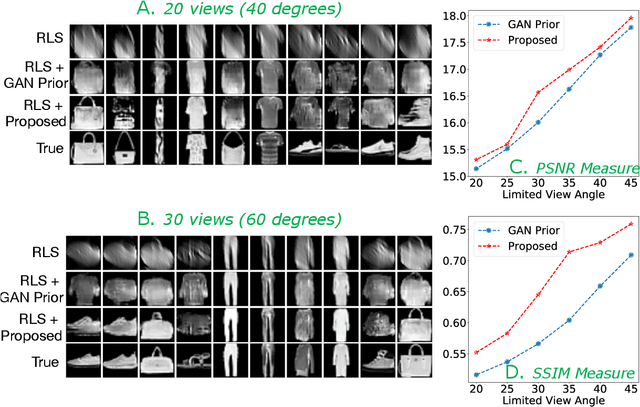
Abstract:Limited angle CT reconstruction is an under-determined linear inverse problem that requires appropriate regularization techniques to be solved. In this work we study how pre-trained generative adversarial networks (GANs) can be used to clean noisy, highly artifact laden reconstructions from conventional techniques, by effectively projecting onto the inferred image manifold. In particular, we use a robust version of the popularly used GAN prior for inverse problems, based on a recent technique called corruption mimicking, that significantly improves the reconstruction quality. The proposed approach operates in the image space directly, as a result of which it does not need to be trained or require access to the measurement model, is scanner agnostic, and can work over a wide range of sensing scenarios.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge