Xiaohua Zhang
Pixel-to-Abundance Translation: Conditional Generative Adversarial Networks Based on Patch Transformer for Hyperspectral Unmixing
Dec 20, 2023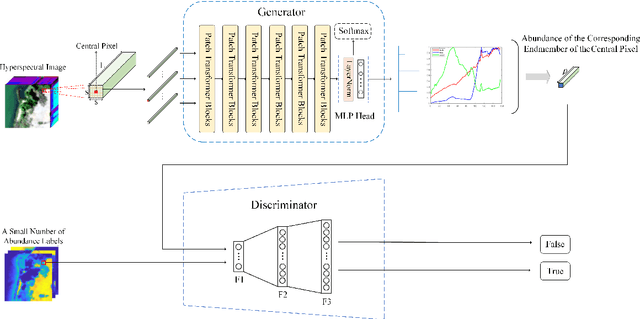
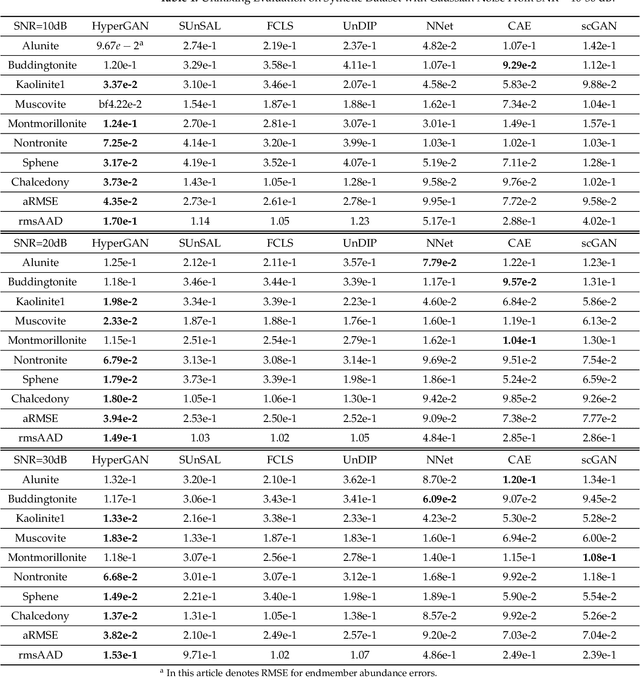
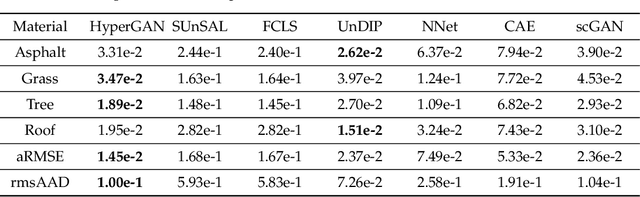
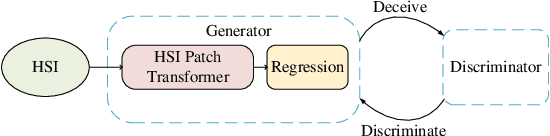
Abstract:Spectral unmixing is a significant challenge in hyperspectral image processing. Existing unmixing methods utilize prior knowledge about the abundance distribution to solve the regularization optimization problem, where the difficulty lies in choosing appropriate prior knowledge and solving the complex regularization optimization problem. To solve these problems, we propose a hyperspectral conditional generative adversarial network (HyperGAN) method as a generic unmixing framework, based on the following assumption: the unmixing process from pixel to abundance can be regarded as a transformation of two modalities with an internal specific relationship. The proposed HyperGAN is composed of a generator and discriminator, the former completes the modal conversion from mixed hyperspectral pixel patch to the abundance of corresponding endmember of the central pixel and the latter is used to distinguish whether the distribution and structure of generated abundance are the same as the true ones. We propose hyperspectral image (HSI) Patch Transformer as the main component of the generator, which utilize adaptive attention score to capture the internal pixels correlation of the HSI patch and leverage the spatial-spectral information in a fine-grained way to achieve optimization of the unmixing process. Experiments on synthetic data and real hyperspectral data achieve impressive results compared to state-of-the-art competitors.
HD-Bind: Encoding of Molecular Structure with Low Precision, Hyperdimensional Binary Representations
Mar 27, 2023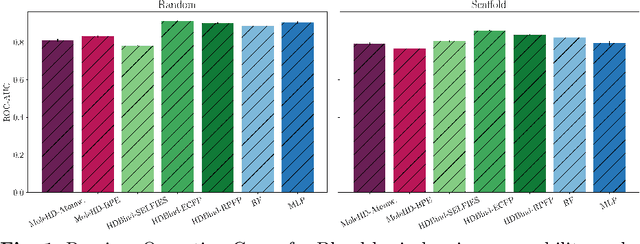
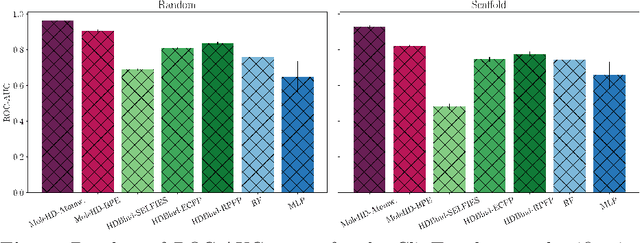
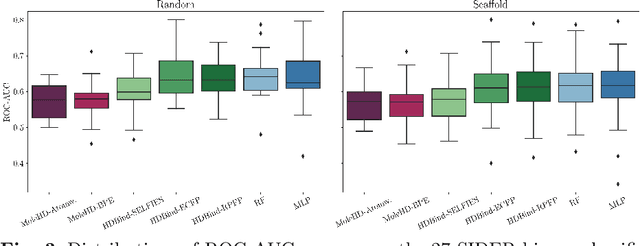
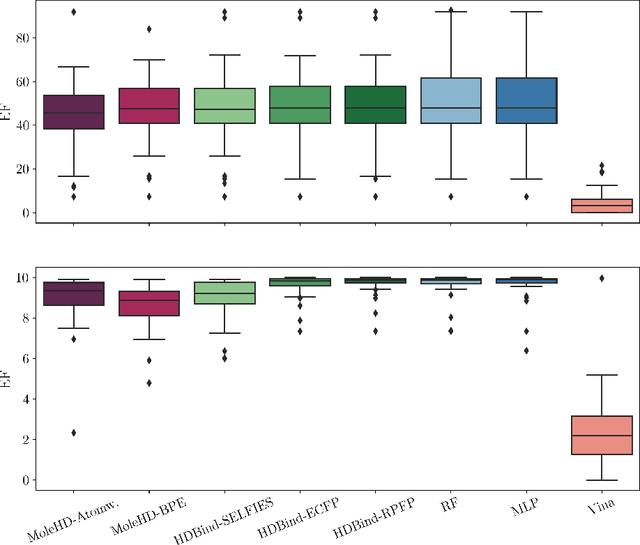
Abstract:Publicly available collections of drug-like molecules have grown to comprise 10s of billions of possibilities in recent history due to advances in chemical synthesis. Traditional methods for identifying ``hit'' molecules from a large collection of potential drug-like candidates have relied on biophysical theory to compute approximations to the Gibbs free energy of the binding interaction between the drug to its protein target. A major drawback of the approaches is that they require exceptional computing capabilities to consider for even relatively small collections of molecules. Hyperdimensional Computing (HDC) is a recently proposed learning paradigm that is able to leverage low-precision binary vector arithmetic to build efficient representations of the data that can be obtained without the need for gradient-based optimization approaches that are required in many conventional machine learning and deep learning approaches. This algorithmic simplicity allows for acceleration in hardware that has been previously demonstrated for a range of application areas. We consider existing HDC approaches for molecular property classification and introduce two novel encoding algorithms that leverage the extended connectivity fingerprint (ECFP) algorithm. We show that HDC-based inference methods are as much as 90 times more efficient than more complex representative machine learning methods and achieve an acceleration of nearly 9 orders of magnitude as compared to inference with molecular docking. We demonstrate multiple approaches for the encoding of molecular data for HDC and examine their relative performance on a range of challenging molecular property prediction and drug-protein binding classification tasks. Our work thus motivates further investigation into molecular representation learning to develop ultra-efficient pre-screening tools.
Evaluating Point-Prediction Uncertainties in Neural Networks for Drug Discovery
Oct 31, 2022



Abstract:Neural Network (NN) models provide potential to speed up the drug discovery process and reduce its failure rates. The success of NN models require uncertainty quantification (UQ) as drug discovery explores chemical space beyond the training data distribution. Standard NN models do not provide uncertainty information. Methods that combine Bayesian models with NN models address this issue, but are difficult to implement and more expensive to train. Some methods require changing the NN architecture or training procedure, limiting the selection of NN models. Moreover, predictive uncertainty can come from different sources. It is important to have the ability to separately model different types of predictive uncertainty, as the model can take assorted actions depending on the source of uncertainty. In this paper, we examine UQ methods that estimate different sources of predictive uncertainty for NN models aiming at drug discovery. We use our prior knowledge on chemical compounds to design the experiments. By utilizing a visualization method we create non-overlapping and chemically diverse partitions from a collection of chemical compounds. These partitions are used as training and test set splits to explore NN model uncertainty. We demonstrate how the uncertainties estimated by the selected methods describe different sources of uncertainty under different partitions and featurization schemes and the relationship to prediction error.
Spatio-Temporal Tuples Transformer for Skeleton-Based Action Recognition
Jan 08, 2022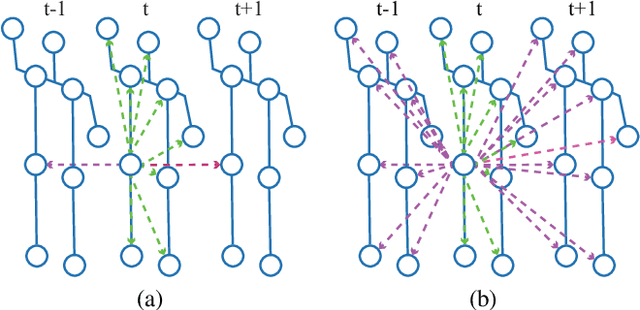
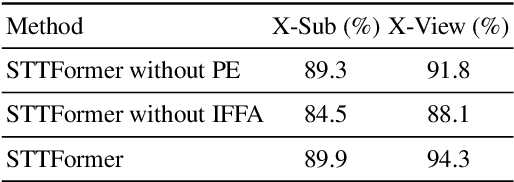
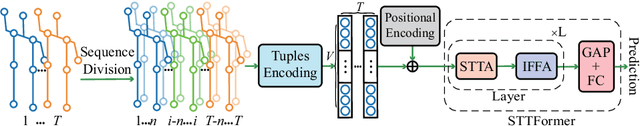
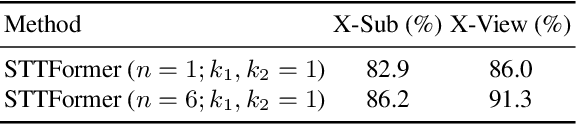
Abstract:Capturing the dependencies between joints is critical in skeleton-based action recognition task. Transformer shows great potential to model the correlation of important joints. However, the existing Transformer-based methods cannot capture the correlation of different joints between frames, which the correlation is very useful since different body parts (such as the arms and legs in "long jump") between adjacent frames move together. Focus on this problem, A novel spatio-temporal tuples Transformer (STTFormer) method is proposed. The skeleton sequence is divided into several parts, and several consecutive frames contained in each part are encoded. And then a spatio-temporal tuples self-attention module is proposed to capture the relationship of different joints in consecutive frames. In addition, a feature aggregation module is introduced between non-adjacent frames to enhance the ability to distinguish similar actions. Compared with the state-of-the-art methods, our method achieves better performance on two large-scale datasets.
High-Throughput Virtual Screening of Small Molecule Inhibitors for SARS-CoV-2 Protein Targets with Deep Fusion Models
Apr 09, 2021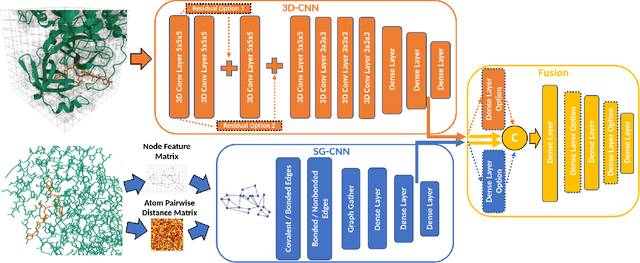
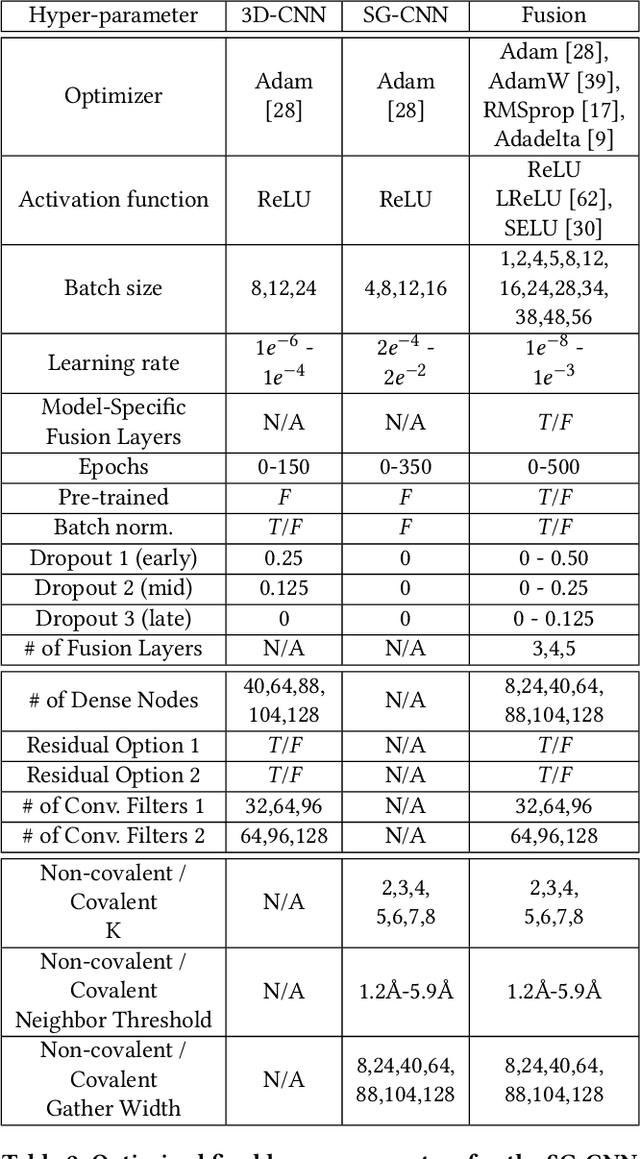
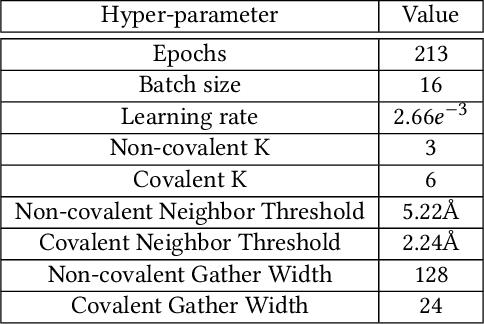
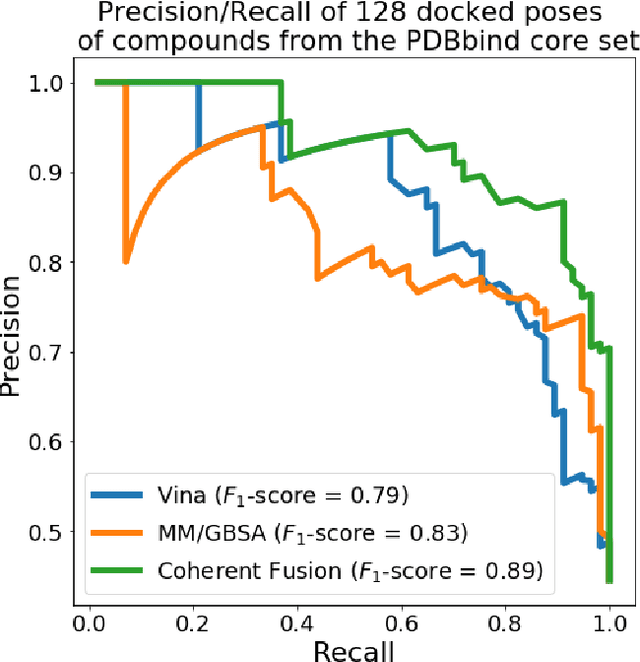
Abstract:Structure-based Deep Fusion models were recently shown to outperform several physics- and machine learning-based protein-ligand binding affinity prediction methods. As part of a multi-institutional COVID-19 pandemic response, over 500 million small molecules were computationally screened against four protein structures from the novel coronavirus (SARS-CoV-2), which causes COVID-19. Three enhancements to Deep Fusion were made in order to evaluate more than 5 billion docked poses on SARS-CoV-2 protein targets. First, the Deep Fusion concept was refined by formulating the architecture as one, coherently backpropagated model (Coherent Fusion) to improve binding-affinity prediction accuracy. Secondly, the model was trained using a distributed, genetic hyper-parameter optimization. Finally, a scalable, high-throughput screening capability was developed to maximize the number of ligands evaluated and expedite the path to experimental evaluation. In this work, we present both the methods developed for machine learning-based high-throughput screening and results from using our computational pipeline to find SARS-CoV-2 inhibitors.
Improved Protein-ligand Binding Affinity Prediction with Structure-Based Deep Fusion Inference
May 17, 2020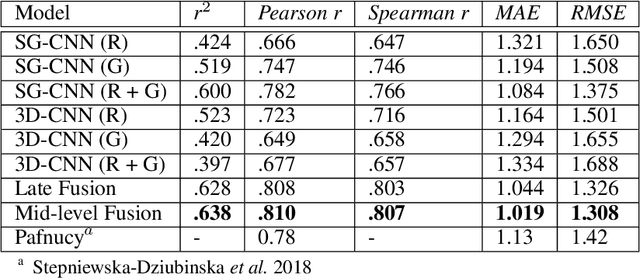
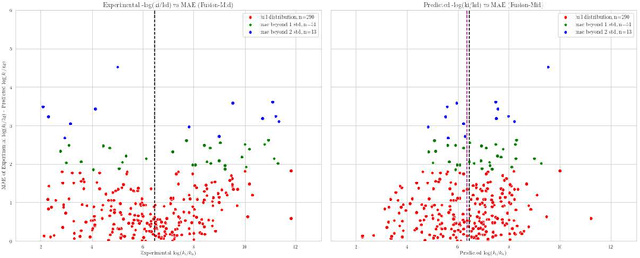
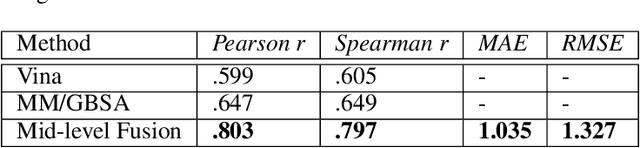
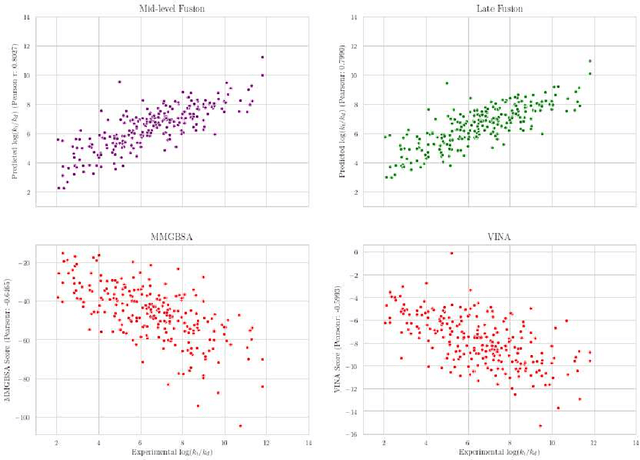
Abstract:Predicting accurate protein-ligand binding affinity is important in drug discovery but remains a challenge even with computationally expensive biophysics-based energy scoring methods and state-of-the-art deep learning approaches. Despite the recent advances in the deep convolutional and graph neural network based approaches, the model performance depends on the input data representation and suffers from distinct limitations. It is natural to combine complementary features and their inference from the individual models for better predictions. We present fusion models to benefit from different feature representations of two neural network models to improve the binding affinity prediction. We demonstrate effectiveness of the proposed approach by performing experiments with the PDBBind 2016 dataset and its docking pose complexes. The results show that the proposed approach improves the overall prediction compared to the individual neural network models with greater computational efficiency than related biophysics based energy scoring functions. We also discuss the benefit of the proposed fusion inference with several example complexes. The software is made available as open source at https://github.com/llnl/fast.
Deep Efficient End-to-end Reconstruction (DEER) Network for Low-dose Few-view Breast CT from Projection Data
Dec 16, 2019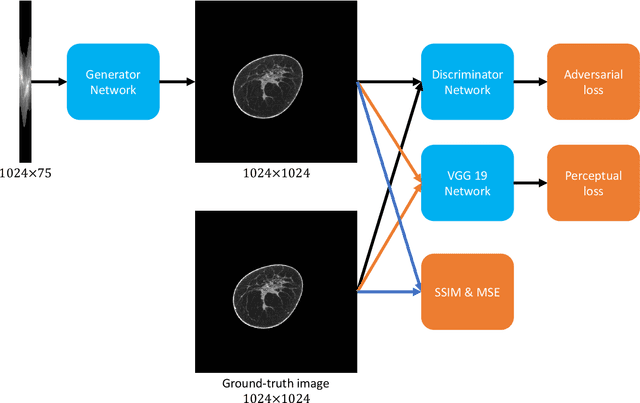
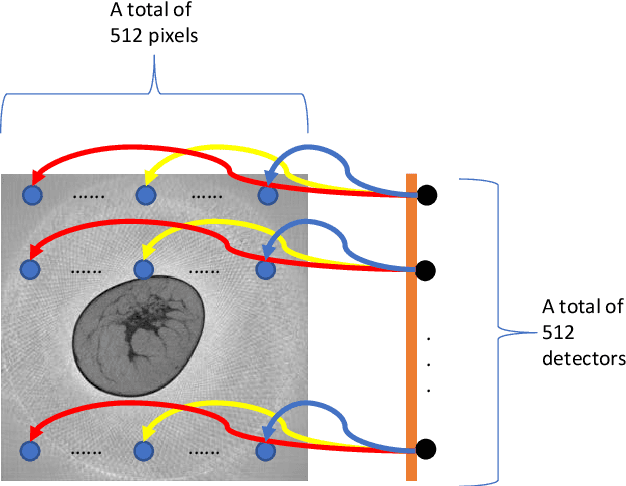
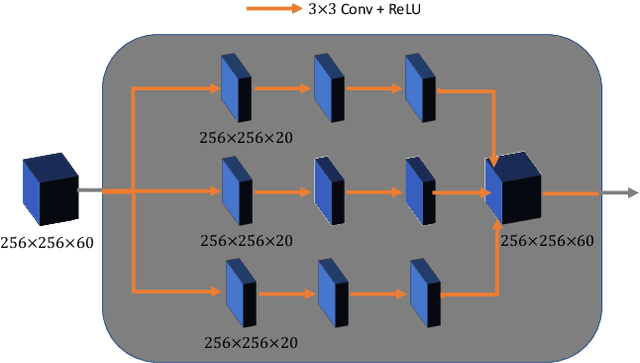
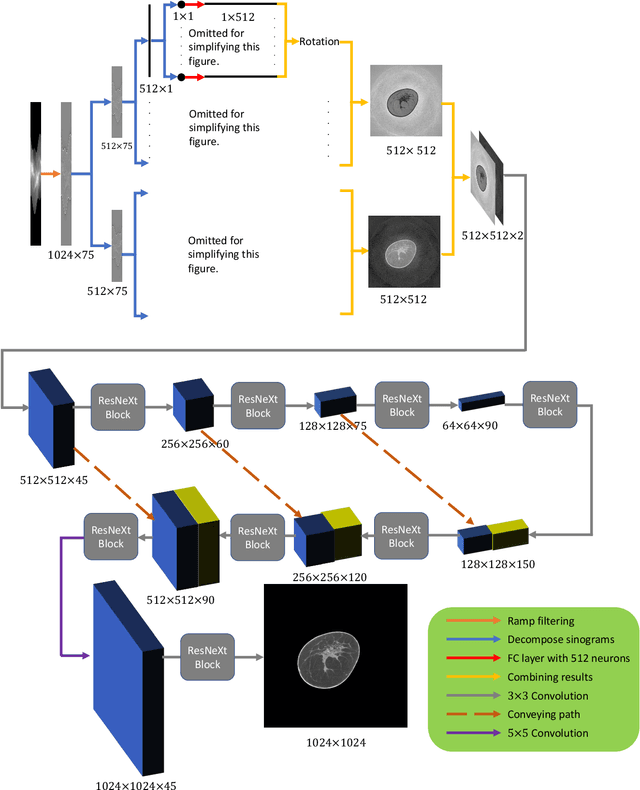
Abstract:Breast CT provides image volumes with isotropic resolution in high contrast, enabling detection of calcification (down to a few hundred microns in size) and subtle density differences. Since breast is sensitive to x-ray radiation, dose reduction of breast CT is an important topic, and for this purpose low-dose few-view scanning is a main approach. In this article, we propose a Deep Efficient End-to-end Reconstruction (DEER) network for low-dose few-view breast CT. The major merits of our network include high dose efficiency, excellent image quality, and low model complexity. By the design, the proposed network can learn the reconstruction process in terms of as less as O(N) parameters, where N is the size of an image to be reconstructed, which represents orders of magnitude improvements relative to the state-of-the-art deep-learning based reconstruction methods that map projection data to tomographic images directly. As a result, our method does not require expensive GPUs to train and run. Also, validated on a cone-beam breast CT dataset prepared by Koning Corporation on a commercial scanner, our method demonstrates competitive performance over the state-of-the-art reconstruction networks in terms of image quality.
Deep-learning-based Breast CT for Radiation Dose Reduction
Sep 25, 2019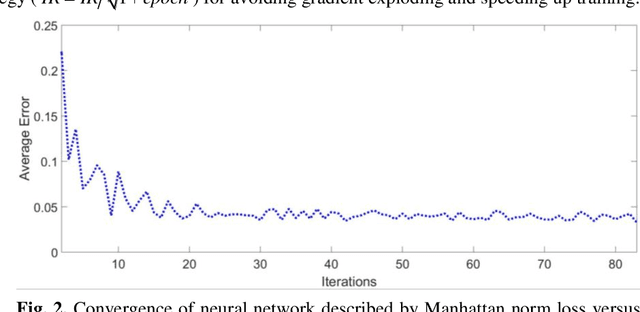
Abstract:Cone-beam breast computed tomography (CT) provides true 3D breast images with isotropic resolution and high-contrast information, detecting calcifications as small as a few hundred microns and revealing subtle tissue differences. However, breast is highly sensitive to x-ray radiation. It is critically important for healthcare to reduce radiation dose. Few-view cone-beam CT only uses a fraction of x-ray projection data acquired by standard cone-beam breast CT, enabling significant reduction of the radiation dose. However, insufficient sampling data would cause severe streak artifacts in CT images reconstructed using conventional methods. In this study, we propose a deep-learning-based method to establish a residual neural network model for the image reconstruction, which is applied for few-view breast CT to produce high quality breast CT images. We respectively evaluate the deep-learning-based image reconstruction using one third and one quarter of x-ray projection views of the standard cone-beam breast CT. Based on clinical breast imaging dataset, we perform a supervised learning to train the neural network from few-view CT images to corresponding full-view CT images. Experimental results show that the deep learning-based image reconstruction method allows few-view breast CT to achieve a radiation dose <6 mGy per cone-beam CT scan, which is a threshold set by FDA for mammographic screening.
Dual Network Architecture for Few-view CT -- Trained on ImageNet Data and Transferred for Medical Imaging
Jul 17, 2019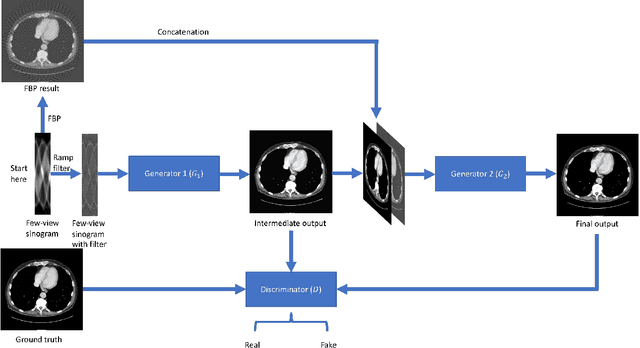
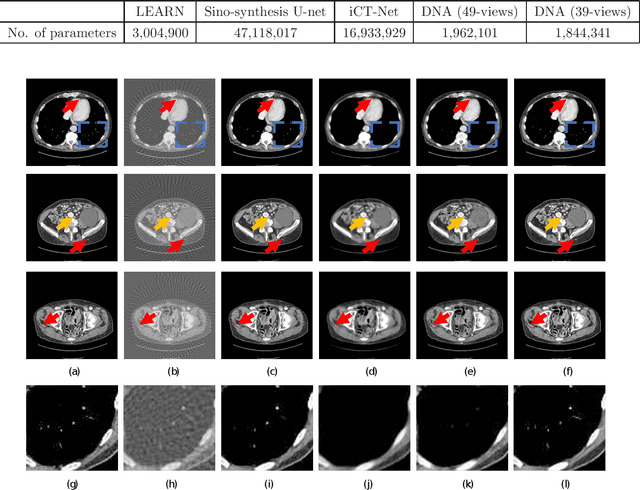
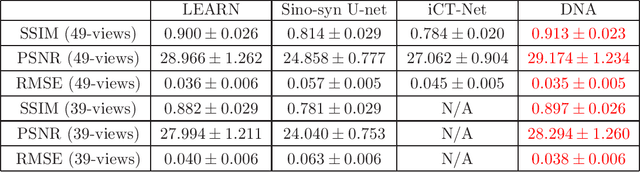
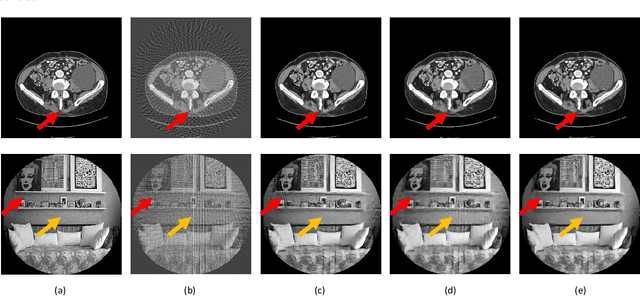
Abstract:X-ray computed tomography (CT) reconstructs cross-sectional images from projection data. However, ionizing X-ray radiation associated with CT scanning might induce cancer and genetic damage and raises public concerns, and the reduction of radiation dose has attracted major attention. Few-view CT image reconstruction is an important topic to reduce the radiation dose. Recently, data-driven algorithms have shown great potential to solve the few-view CT problem. In this paper, we develop a dual network architecture (DNA) for reconstructing images directly from sinograms. In the proposed DNA method, a point-based fully-connected layer learns the backprojection process requesting significantly less memory than the prior art and with O(C*N*N_c) parameters where N and N_c denote the dimension of reconstructed images and number of projections respectively. C is an adjustable parameter that can be set as low as 1. Our experimental results demonstrate that DNA produces a competitive performance over the other state-of-the-art methods. Interestingly, natural images can be used to pre-train DNA to avoid overfitting when the amount of real patient images is limited.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge