Weihong Xu
Lab of Efficient Architectures for Digital-communication and Signal-processing, National Mobile Communications Research Laboratory
Illusions of Confidence? Diagnosing LLM Truthfulness via Neighborhood Consistency
Jan 09, 2026Abstract:As Large Language Models (LLMs) are increasingly deployed in real-world settings, correctness alone is insufficient. Reliable deployment requires maintaining truthful beliefs under contextual perturbations. Existing evaluations largely rely on point-wise confidence like Self-Consistency, which can mask brittle belief. We show that even facts answered with perfect self-consistency can rapidly collapse under mild contextual interference. To address this gap, we propose Neighbor-Consistency Belief (NCB), a structural measure of belief robustness that evaluates response coherence across a conceptual neighborhood. To validate the efficiency of NCB, we introduce a new cognitive stress-testing protocol that probes outputs stability under contextual interference. Experiments across multiple LLMs show that the performance of high-NCB data is relatively more resistant to interference. Finally, we present Structure-Aware Training (SAT), which optimizes context-invariant belief structure and reduces long-tail knowledge brittleness by approximately 30%. Code will be available at https://github.com/zjunlp/belief.
Clo-HDnn: A 4.66 TFLOPS/W and 3.78 TOPS/W Continual On-Device Learning Accelerator with Energy-efficient Hyperdimensional Computing via Progressive Search
Jul 23, 2025Abstract:Clo-HDnn is an on-device learning (ODL) accelerator designed for emerging continual learning (CL) tasks. Clo-HDnn integrates hyperdimensional computing (HDC) along with low-cost Kronecker HD Encoder and weight clustering feature extraction (WCFE) to optimize accuracy and efficiency. Clo-HDnn adopts gradient-free CL to efficiently update and store the learned knowledge in the form of class hypervectors. Its dual-mode operation enables bypassing costly feature extraction for simpler datasets, while progressive search reduces complexity by up to 61% by encoding and comparing only partial query hypervectors. Achieving 4.66 TFLOPS/W (FE) and 3.78 TOPS/W (classifier), Clo-HDnn delivers 7.77x and 4.85x higher energy efficiency compared to SOTA ODL accelerators.
FSL-HDnn: A 5.7 TOPS/W End-to-end Few-shot Learning Classifier Accelerator with Feature Extraction and Hyperdimensional Computing
Sep 17, 2024



Abstract:This paper introduces FSL-HDnn, an energy-efficient accelerator that implements the end-to-end pipeline of feature extraction, classification, and on-chip few-shot learning (FSL) through gradient-free learning techniques in a 40 nm CMOS process. At its core, FSL-HDnn integrates two low-power modules: Weight clustering feature extractor and Hyperdimensional Computing (HDC). Feature extractor utilizes advanced weight clustering and pattern reuse strategies for optimized CNN-based feature extraction. Meanwhile, HDC emerges as a novel approach for lightweight FSL classifier, employing hyperdimensional vectors to improve training accuracy significantly compared to traditional distance-based approaches. This dual-module synergy not only simplifies the learning process by eliminating the need for complex gradients but also dramatically enhances energy efficiency and performance. Specifically, FSL-HDnn achieves an Intensity unprecedented energy efficiency of 5.7 TOPS/W for feature 1 extraction and 0.78 TOPS/W for classification and learning Training Intensity phases, achieving improvements of 2.6X and 6.6X, respectively, Storage over current state-of-the-art CNN and FSL processors.
SpecHD: Hyperdimensional Computing Framework for FPGA-based Mass Spectrometry Clustering
Nov 20, 2023Abstract:Mass spectrometry-based proteomics is a key enabler for personalized healthcare, providing a deep dive into the complex protein compositions of biological systems. This technology has vast applications in biotechnology and biomedicine but faces significant computational bottlenecks. Current methodologies often require multiple hours or even days to process extensive datasets, particularly in the domain of spectral clustering. To tackle these inefficiencies, we introduce SpecHD, a hyperdimensional computing (HDC) framework supplemented by an FPGA-accelerated architecture with integrated near-storage preprocessing. Utilizing streamlined binary operations in an HDC environment, SpecHD capitalizes on the low-latency and parallel capabilities of FPGAs. This approach markedly improves clustering speed and efficiency, serving as a catalyst for real-time, high-throughput data analysis in future healthcare applications. Our evaluations demonstrate that SpecHD not only maintains but often surpasses existing clustering quality metrics while drastically cutting computational time. Specifically, it can cluster a large-scale human proteome dataset-comprising 25 million MS/MS spectra and 131 GB of MS data-in just 5 minutes. With energy efficiency exceeding 31x and a speedup factor that spans a range of 6x to 54x over existing state of-the-art solutions, SpecHD emerges as a promising solution for the rapid analysis of mass spectrometry data with great implications for personalized healthcare.
HD-Bind: Encoding of Molecular Structure with Low Precision, Hyperdimensional Binary Representations
Mar 27, 2023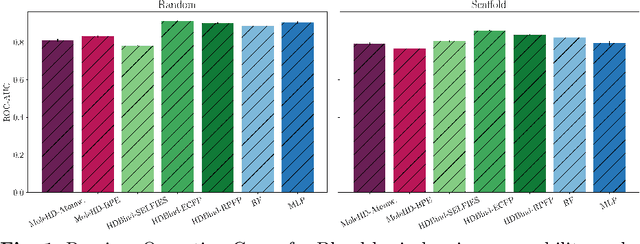
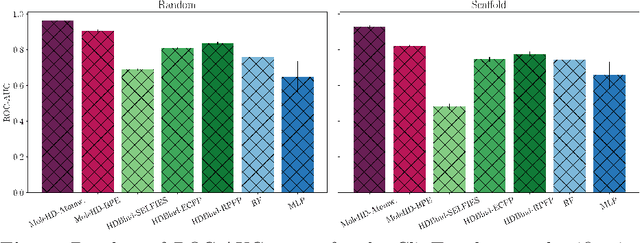
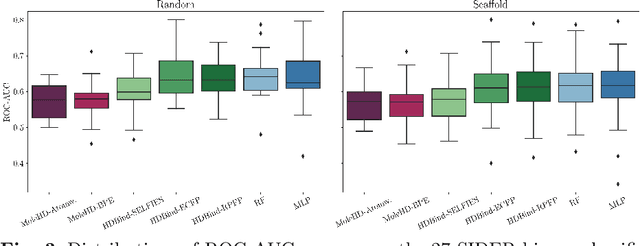
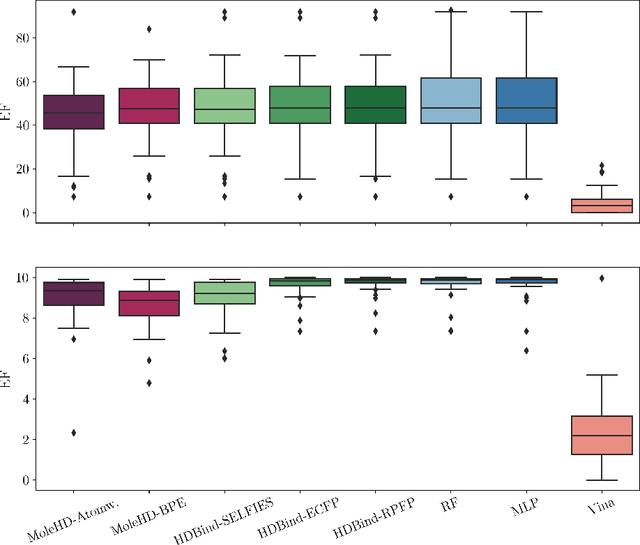
Abstract:Publicly available collections of drug-like molecules have grown to comprise 10s of billions of possibilities in recent history due to advances in chemical synthesis. Traditional methods for identifying ``hit'' molecules from a large collection of potential drug-like candidates have relied on biophysical theory to compute approximations to the Gibbs free energy of the binding interaction between the drug to its protein target. A major drawback of the approaches is that they require exceptional computing capabilities to consider for even relatively small collections of molecules. Hyperdimensional Computing (HDC) is a recently proposed learning paradigm that is able to leverage low-precision binary vector arithmetic to build efficient representations of the data that can be obtained without the need for gradient-based optimization approaches that are required in many conventional machine learning and deep learning approaches. This algorithmic simplicity allows for acceleration in hardware that has been previously demonstrated for a range of application areas. We consider existing HDC approaches for molecular property classification and introduce two novel encoding algorithms that leverage the extended connectivity fingerprint (ECFP) algorithm. We show that HDC-based inference methods are as much as 90 times more efficient than more complex representative machine learning methods and achieve an acceleration of nearly 9 orders of magnitude as compared to inference with molecular docking. We demonstrate multiple approaches for the encoding of molecular data for HDC and examine their relative performance on a range of challenging molecular property prediction and drug-protein binding classification tasks. Our work thus motivates further investigation into molecular representation learning to develop ultra-efficient pre-screening tools.
Polar Decoding on Sparse Graphs with Deep Learning
Nov 24, 2018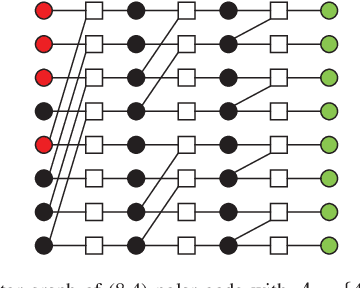
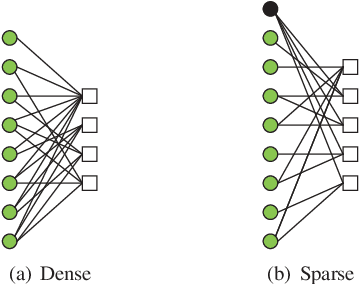
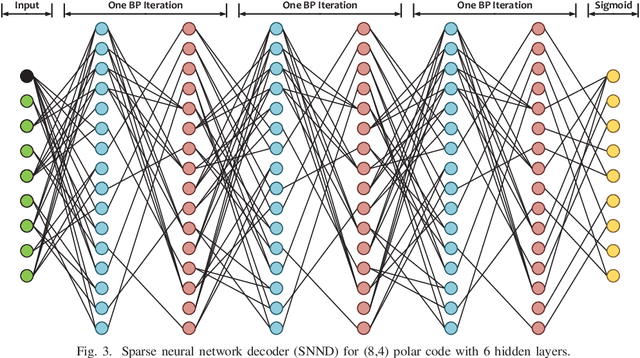
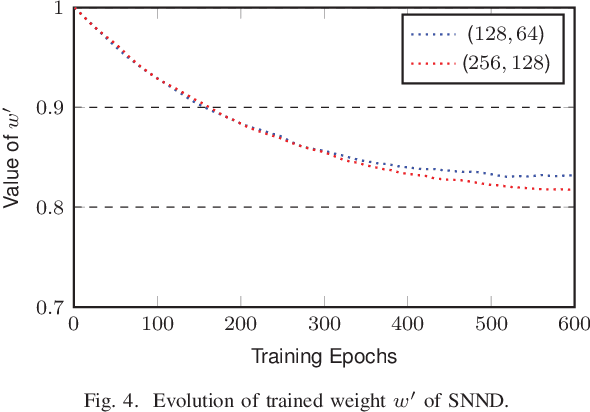
Abstract:In this paper, we present a sparse neural network decoder (SNND) of polar codes based on belief propagation (BP) and deep learning. At first, the conventional factor graph of polar BP decoding is converted to the bipartite Tanner graph similar to low-density parity-check (LDPC) codes. Then the Tanner graph is unfolded and translated into the graphical representation of deep neural network (DNN). The complex sum-product algorithm (SPA) is modified to min-sum (MS) approximation with low complexity. We dramatically reduce the number of weight by using single weight to parameterize the networks. Optimized by the training techniques of deep learning, proposed SNND achieves comparative decoding performance of SPA and obtains about $0.5$ dB gain over MS decoding on ($128,64$) and ($256,128$) codes. Moreover, $60 \%$ complexity reduction is achieved and the decoding latency is significantly lower than the conventional polar BP.
Improving Massive MIMO Belief Propagation Detector with Deep Neural Network
Apr 05, 2018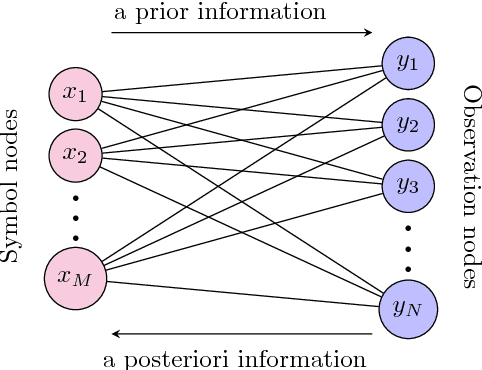
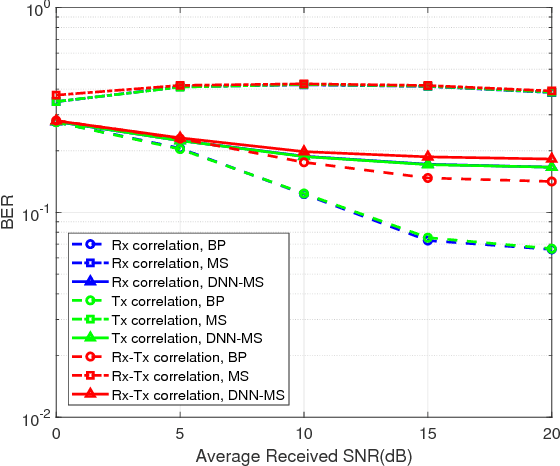
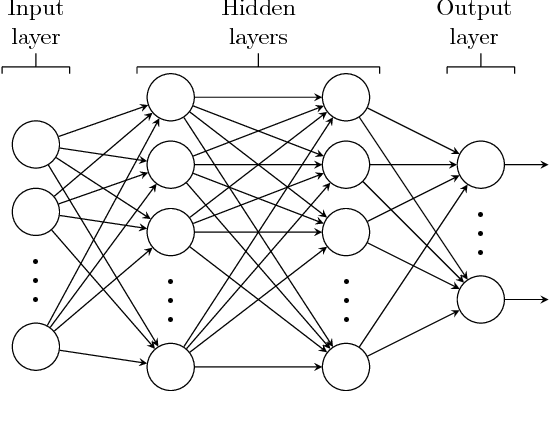
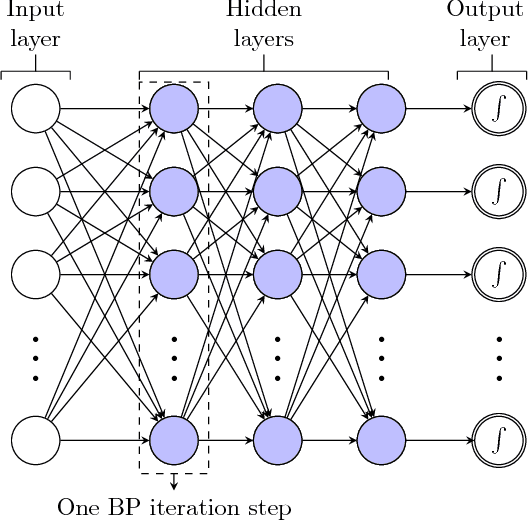
Abstract:In this paper, deep neural network (DNN) is utilized to improve the belief propagation (BP) detection for massive multiple-input multiple-output (MIMO) systems. A neural network architecture suitable for detection task is firstly introduced by unfolding BP algorithms. DNN MIMO detectors are then proposed based on two modified BP detectors, damped BP and max-sum BP. The correction factors in these algorithms are optimized through deep learning techniques, aiming at improved detection performance. Numerical results are presented to demonstrate the performance of the DNN detectors in comparison with various BP modifications. The neural network is trained once and can be used for multiple online detections. The results show that, compared to other state-of-the-art detectors, the DNN detectors can achieve lower bit error rate (BER) with improved robustness against various antenna configurations and channel conditions at the same level of complexity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge