Garrett A. Stevenson
Distributed Stochastic Optimization of a Neural Representation Network for Time-Space Tomography Reconstruction
Apr 29, 2024



Abstract:4D time-space reconstruction of dynamic events or deforming objects using X-ray computed tomography (CT) is an extremely ill-posed inverse problem. Existing approaches assume that the object remains static for the duration of several tens or hundreds of X-ray projection measurement images (reconstruction of consecutive limited-angle CT scans). However, this is an unrealistic assumption for many in-situ experiments that causes spurious artifacts and inaccurate morphological reconstructions of the object. To solve this problem, we propose to perform a 4D time-space reconstruction using a distributed implicit neural representation (DINR) network that is trained using a novel distributed stochastic training algorithm. Our DINR network learns to reconstruct the object at its output by iterative optimization of its network parameters such that the measured projection images best match the output of the CT forward measurement model. We use a continuous time and space forward measurement model that is a function of the DINR outputs at a sparsely sampled set of continuous valued object coordinates. Unlike existing state-of-the-art neural representation architectures that forward and back propagate through dense voxel grids that sample the object's entire time-space coordinates, we only propagate through the DINR at a small subset of object coordinates in each iteration resulting in an order-of-magnitude reduction in memory and compute for training. DINR leverages distributed computation across several compute nodes and GPUs to produce high-fidelity 4D time-space reconstructions even for extremely large CT data sizes. We use both simulated parallel-beam and experimental cone-beam X-ray CT datasets to demonstrate the superior performance of our approach.
High-Throughput Virtual Screening of Small Molecule Inhibitors for SARS-CoV-2 Protein Targets with Deep Fusion Models
Apr 09, 2021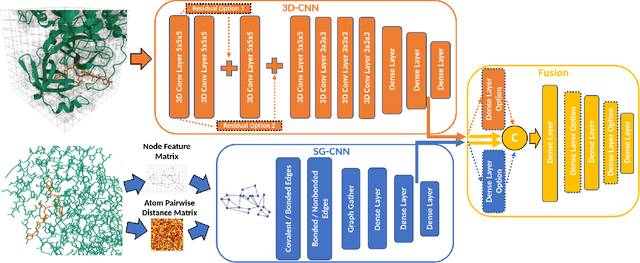
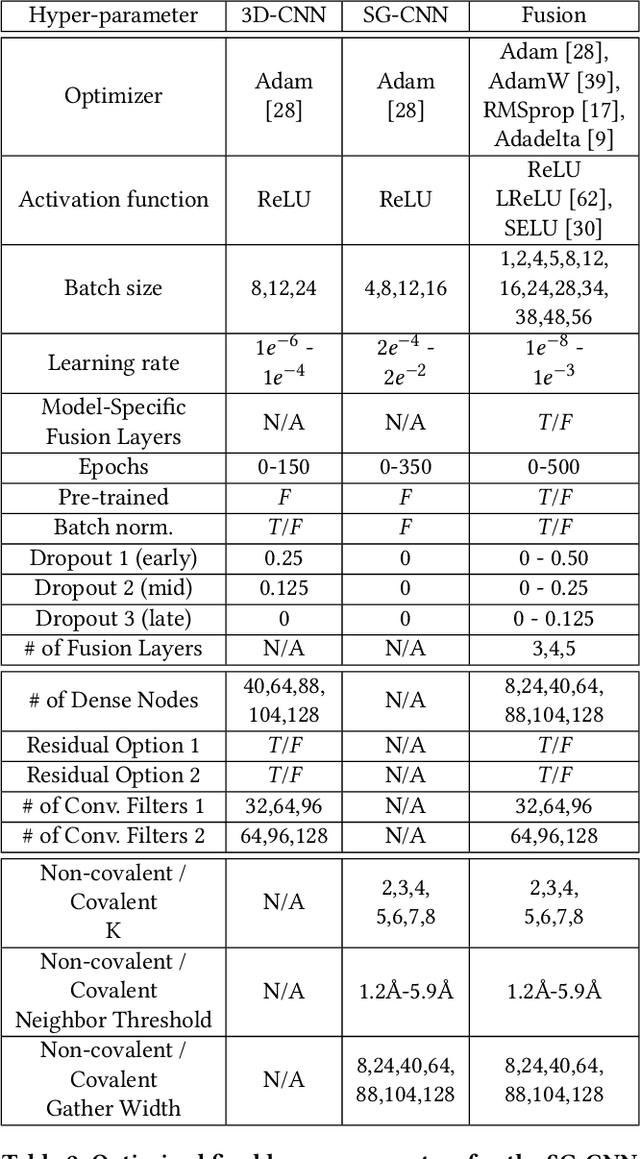
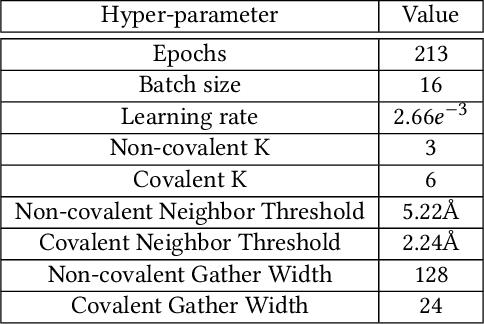
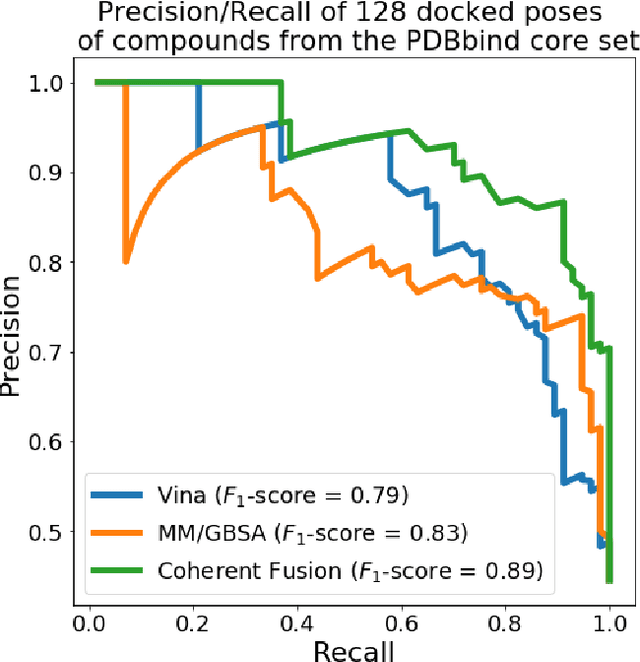
Abstract:Structure-based Deep Fusion models were recently shown to outperform several physics- and machine learning-based protein-ligand binding affinity prediction methods. As part of a multi-institutional COVID-19 pandemic response, over 500 million small molecules were computationally screened against four protein structures from the novel coronavirus (SARS-CoV-2), which causes COVID-19. Three enhancements to Deep Fusion were made in order to evaluate more than 5 billion docked poses on SARS-CoV-2 protein targets. First, the Deep Fusion concept was refined by formulating the architecture as one, coherently backpropagated model (Coherent Fusion) to improve binding-affinity prediction accuracy. Secondly, the model was trained using a distributed, genetic hyper-parameter optimization. Finally, a scalable, high-throughput screening capability was developed to maximize the number of ligands evaluated and expedite the path to experimental evaluation. In this work, we present both the methods developed for machine learning-based high-throughput screening and results from using our computational pipeline to find SARS-CoV-2 inhibitors.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge