Hans-Peter Seidel
Max Planck Institute for Informatics, Saarland Informatics Campus
Uncertainty separation via ensemble quantile regression
Dec 18, 2024



Abstract:This paper introduces a novel and scalable framework for uncertainty estimation and separation with applications in data driven modeling in science and engineering tasks where reliable uncertainty quantification is critical. Leveraging an ensemble of quantile regression (E-QR) models, our approach enhances aleatoric uncertainty estimation while preserving the quality of epistemic uncertainty, surpassing competing methods, such as Deep Ensembles (DE) and Monte Carlo (MC) dropout. To address challenges in separating uncertainty types, we propose an algorithm that iteratively improves separation through progressive sampling in regions of high uncertainty. Our framework is scalable to large datasets and demonstrates superior performance on synthetic benchmarks, offering a robust tool for uncertainty quantification in data-driven applications.
Two-Stage Pretraining for Molecular Property Prediction in the Wild
Nov 05, 2024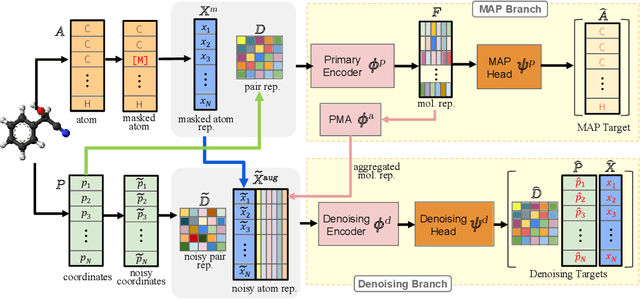

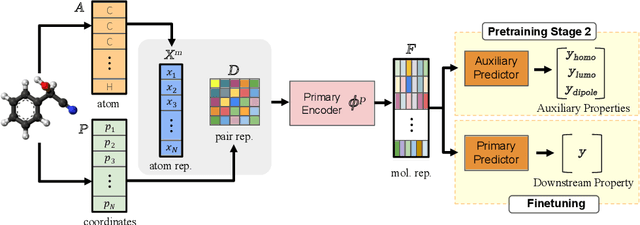
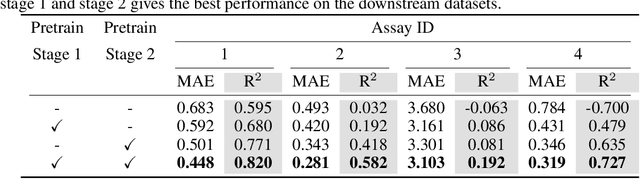
Abstract:Accurate property prediction is crucial for accelerating the discovery of new molecules. Although deep learning models have achieved remarkable success, their performance often relies on large amounts of labeled data that are expensive and time-consuming to obtain. Thus, there is a growing need for models that can perform well with limited experimentally-validated data. In this work, we introduce MoleVers, a versatile pretrained model designed for various types of molecular property prediction in the wild, i.e., where experimentally-validated molecular property labels are scarce. MoleVers adopts a two-stage pretraining strategy. In the first stage, the model learns molecular representations from large unlabeled datasets via masked atom prediction and dynamic denoising, a novel task enabled by a new branching encoder architecture. In the second stage, MoleVers is further pretrained using auxiliary labels obtained with inexpensive computational methods, enabling supervised learning without the need for costly experimental data. This two-stage framework allows MoleVers to learn representations that generalize effectively across various downstream datasets. We evaluate MoleVers on a new benchmark comprising 22 molecular datasets with diverse types of properties, the majority of which contain 50 or fewer training labels reflecting real-world conditions. MoleVers achieves state-of-the-art results on 20 out of the 22 datasets, and ranks second among the remaining two, highlighting its ability to bridge the gap between data-hungry models and real-world conditions where practically-useful labels are scarce.
Learning Images Across Scales Using Adversarial Training
Jun 13, 2024



Abstract:The real world exhibits rich structure and detail across many scales of observation. It is difficult, however, to capture and represent a broad spectrum of scales using ordinary images. We devise a novel paradigm for learning a representation that captures an orders-of-magnitude variety of scales from an unstructured collection of ordinary images. We treat this collection as a distribution of scale-space slices to be learned using adversarial training, and additionally enforce coherency across slices. Our approach relies on a multiscale generator with carefully injected procedural frequency content, which allows to interactively explore the emerging continuous scale space. Training across vastly different scales poses challenges regarding stability, which we tackle using a supervision scheme that involves careful sampling of scales. We show that our generator can be used as a multiscale generative model, and for reconstructions of scale spaces from unstructured patches. Significantly outperforming the state of the art, we demonstrate zoom-in factors of up to 256x at high quality and scale consistency.
Cinematic Gaussians: Real-Time HDR Radiance Fields with Depth of Field
Jun 11, 2024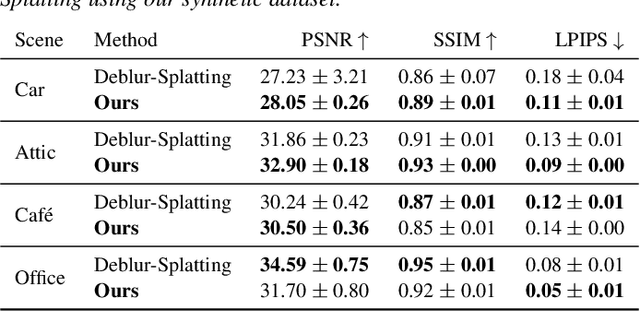
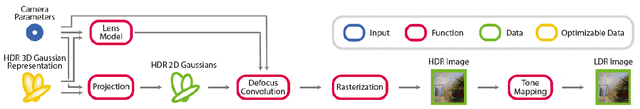
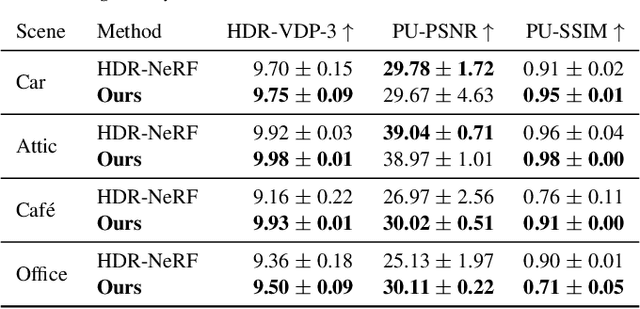

Abstract:Radiance field methods represent the state of the art in reconstructing complex scenes from multi-view photos. However, these reconstructions often suffer from one or both of the following limitations: First, they typically represent scenes in low dynamic range (LDR), which restricts their use to evenly lit environments and hinders immersive viewing experiences. Secondly, their reliance on a pinhole camera model, assuming all scene elements are in focus in the input images, presents practical challenges and complicates refocusing during novel-view synthesis. Addressing these limitations, we present a lightweight method based on 3D Gaussian Splatting that utilizes multi-view LDR images of a scene with varying exposure times, apertures, and focus distances as input to reconstruct a high-dynamic-range (HDR) radiance field. By incorporating analytical convolutions of Gaussians based on a thin-lens camera model as well as a tonemapping module, our reconstructions enable the rendering of HDR content with flexible refocusing capabilities. We demonstrate that our combined treatment of HDR and depth of field facilitates real-time cinematic rendering, outperforming the state of the art.
Neural Gaussian Scale-Space Fields
May 31, 2024



Abstract:Gaussian scale spaces are a cornerstone of signal representation and processing, with applications in filtering, multiscale analysis, anti-aliasing, and many more. However, obtaining such a scale space is costly and cumbersome, in particular for continuous representations such as neural fields. We present an efficient and lightweight method to learn the fully continuous, anisotropic Gaussian scale space of an arbitrary signal. Based on Fourier feature modulation and Lipschitz bounding, our approach is trained self-supervised, i.e., training does not require any manual filtering. Our neural Gaussian scale-space fields faithfully capture multiscale representations across a broad range of modalities, and support a diverse set of applications. These include images, geometry, light-stage data, texture anti-aliasing, and multiscale optimization.
Exposure Diffusion: HDR Image Generation by Consistent LDR denoising
May 23, 2024

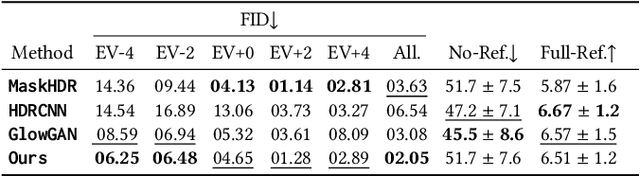
Abstract:We demonstrate generating high-dynamic range (HDR) images using the concerted action of multiple black-box, pre-trained low-dynamic range (LDR) image diffusion models. Common diffusion models are not HDR as, first, there is no sufficiently large HDR image dataset available to re-train them, and second, even if it was, re-training such models is impossible for most compute budgets. Instead, we seek inspiration from the HDR image capture literature that traditionally fuses sets of LDR images, called "brackets", to produce a single HDR image. We operate multiple denoising processes to generate multiple LDR brackets that together form a valid HDR result. To this end, we introduce an exposure consistency term into the diffusion process to couple the brackets such that they agree across the exposure range they share. We demonstrate HDR versions of state-of-the-art unconditional and conditional as well as restoration-type (LDR2HDR) generative modeling.
TrustMol: Trustworthy Inverse Molecular Design via Alignment with Molecular Dynamics
Feb 26, 2024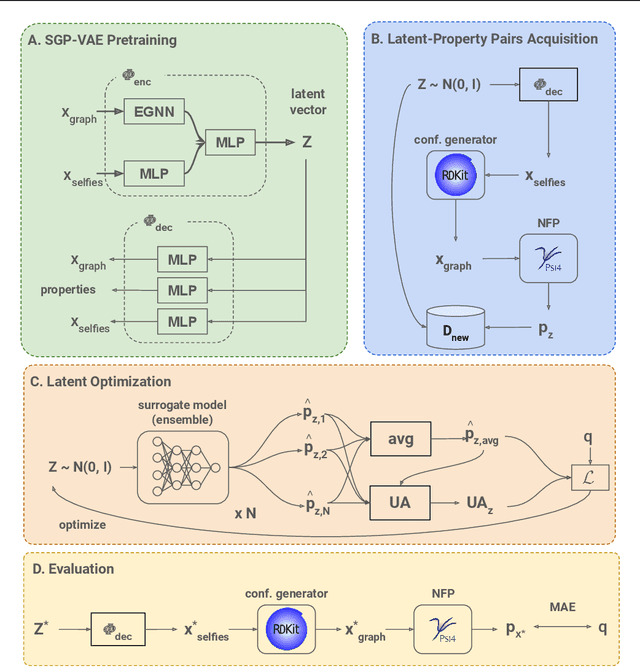
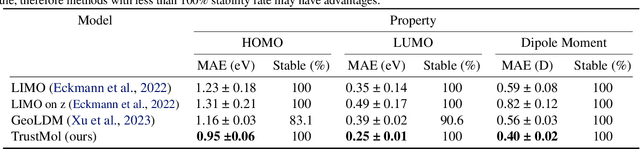
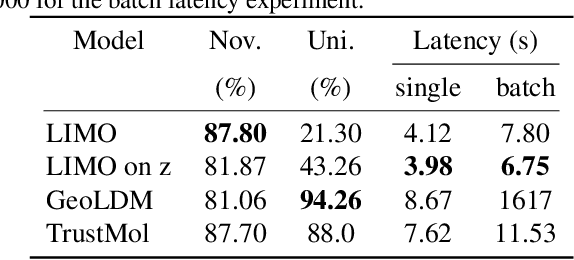
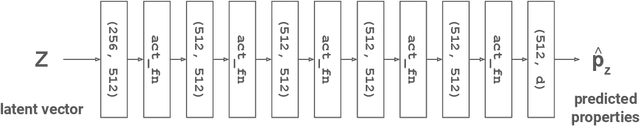
Abstract:Data-driven generation of molecules with desired properties, also known as inverse molecular design (IMD), has attracted significant attention in recent years. Despite the significant progress in the accuracy and diversity of solutions, existing IMD methods lag behind in terms of trustworthiness. The root issue is that the design process of these methods is increasingly more implicit and indirect, and this process is also isolated from the native forward process (NFP), the ground-truth function that models the molecular dynamics. Following this insight, we propose TrustMol, an IMD method built to be trustworthy. For this purpose, TrustMol relies on a set of technical novelties including a new variational autoencoder network. Moreover, we propose a latent-property pairs acquisition method to effectively navigate the complexities of molecular latent optimization, a process that seems intuitive yet challenging due to the high-frequency and discontinuous nature of molecule space. TrustMol also integrates uncertainty-awareness into molecular latent optimization. These lead to improvements in both explainability and reliability of the IMD process. We validate the trustworthiness of TrustMol through a wide range of experiments.
Joint Sampling and Optimisation for Inverse Rendering
Sep 27, 2023



Abstract:When dealing with difficult inverse problems such as inverse rendering, using Monte Carlo estimated gradients to optimise parameters can slow down convergence due to variance. Averaging many gradient samples in each iteration reduces this variance trivially. However, for problems that require thousands of optimisation iterations, the computational cost of this approach rises quickly. We derive a theoretical framework for interleaving sampling and optimisation. We update and reuse past samples with low-variance finite-difference estimators that describe the change in the estimated gradients between each iteration. By combining proportional and finite-difference samples, we continuously reduce the variance of our novel gradient meta-estimators throughout the optimisation process. We investigate how our estimator interlinks with Adam and derive a stable combination. We implement our method for inverse path tracing and demonstrate how our estimator speeds up convergence on difficult optimisation tasks.
Large-Batch, Neural Multi-Objective Bayesian Optimization
Jun 12, 2023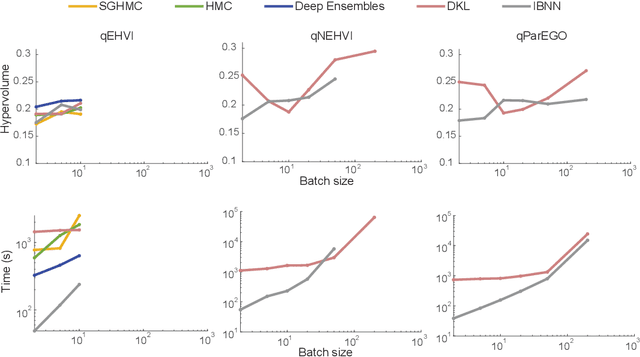
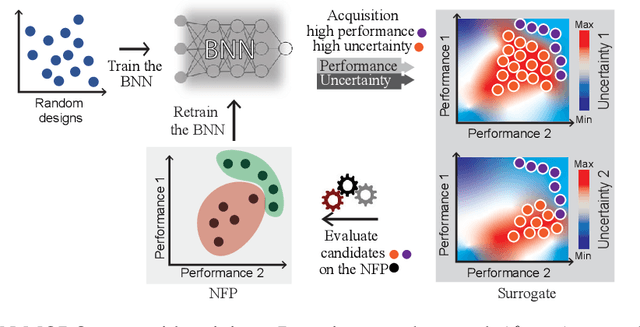
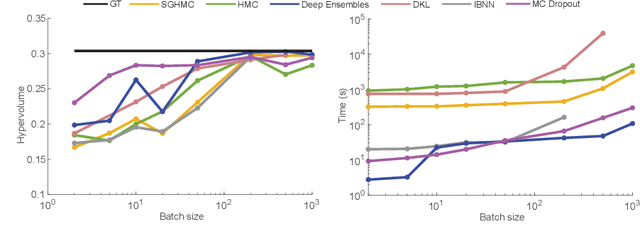
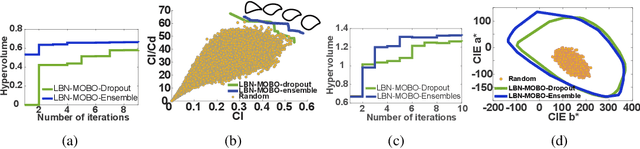
Abstract:Bayesian optimization provides a powerful framework for global optimization of black-box, expensive-to-evaluate functions. However, it has a limited capacity in handling data-intensive problems, especially in multi-objective settings, due to the poor scalability of default Gaussian Process surrogates. We present a novel Bayesian optimization framework specifically tailored to address these limitations. Our method leverages a Bayesian neural networks approach for surrogate modeling. This enables efficient handling of large batches of data, modeling complex problems, and generating the uncertainty of the predictions. In addition, our method incorporates a scalable, uncertainty-aware acquisition strategy based on the well-known, easy-to-deploy NSGA-II. This fully parallelizable strategy promotes efficient exploration of uncharted regions. Our framework allows for effective optimization in data-intensive environments with a minimum number of iterations. We demonstrate the superiority of our method by comparing it with state-of-the-art multi-objective optimizations. We perform our evaluation on two real-world problems - airfoil design and color printing - showcasing the applicability and efficiency of our approach. Code is available at: https://github.com/an-on-ym-ous/lbn_mobo
Enhancing image quality prediction with self-supervised visual masking
May 31, 2023Abstract:Full-reference image quality metrics (FR-IQMs) aim to measure the visual differences between a pair of reference and distorted images, with the goal of accurately predicting human judgments. However, existing FR-IQMs, including traditional ones like PSNR and SSIM and even perceptual ones such as HDR-VDP, LPIPS, and DISTS, still fall short in capturing the complexities and nuances of human perception. In this work, rather than devising a novel IQM model, we seek to improve upon the perceptual quality of existing FR-IQM methods. We achieve this by considering visual masking, an important characteristic of the human visual system that changes its sensitivity to distortions as a function of local image content. Specifically, for a given FR-IQM metric, we propose to predict a visual masking model that modulates reference and distorted images in a way that penalizes the visual errors based on their visibility. Since the ground truth visual masks are difficult to obtain, we demonstrate how they can be derived in a self-supervised manner solely based on mean opinion scores (MOS) collected from an FR-IQM dataset. Our approach results in enhanced FR-IQM metrics that are more in line with human prediction both visually and quantitatively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge