Gustaf Ahdritz
Provable Uncertainty Decomposition via Higher-Order Calibration
Dec 25, 2024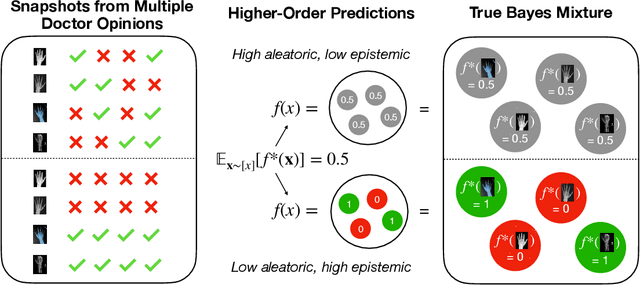
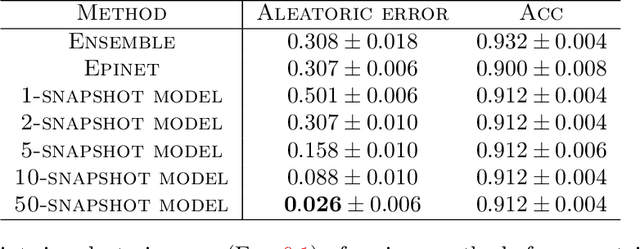
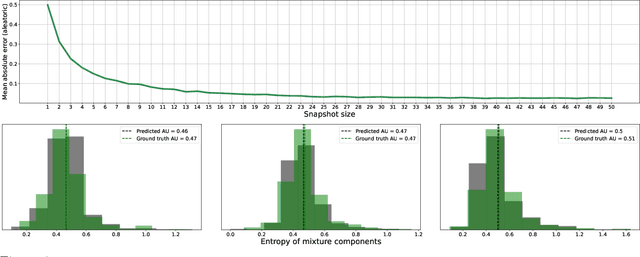
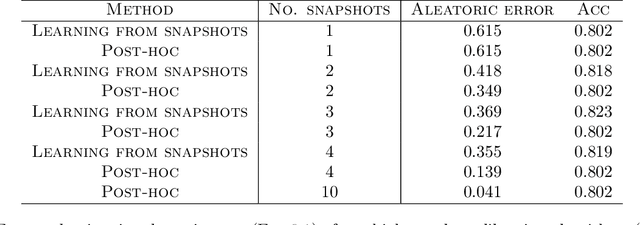
Abstract:We give a principled method for decomposing the predictive uncertainty of a model into aleatoric and epistemic components with explicit semantics relating them to the real-world data distribution. While many works in the literature have proposed such decompositions, they lack the type of formal guarantees we provide. Our method is based on the new notion of higher-order calibration, which generalizes ordinary calibration to the setting of higher-order predictors that predict mixtures over label distributions at every point. We show how to measure as well as achieve higher-order calibration using access to $k$-snapshots, namely examples where each point has $k$ independent conditional labels. Under higher-order calibration, the estimated aleatoric uncertainty at a point is guaranteed to match the real-world aleatoric uncertainty averaged over all points where the prediction is made. To our knowledge, this is the first formal guarantee of this type that places no assumptions whatsoever on the real-world data distribution. Importantly, higher-order calibration is also applicable to existing higher-order predictors such as Bayesian and ensemble models and provides a natural evaluation metric for such models. We demonstrate through experiments that our method produces meaningful uncertainty decompositions for image classification.
Modeling Real-Time Interactive Conversations as Timed Diarized Transcripts
May 21, 2024



Abstract:Chatbots built upon language models have exploded in popularity, but they have largely been limited to synchronous, turn-by-turn dialogues. In this paper we present a simple yet general method to simulate real-time interactive conversations using pretrained text-only language models, by modeling timed diarized transcripts and decoding them with causal rejection sampling. We demonstrate the promise of this method with two case studies: instant messenger dialogues and spoken conversations, which require generation at about 30 tok/s and 20 tok/s respectively to maintain real-time interactivity. These capabilities can be added into language models using relatively little data and run on commodity hardware.
Distinguishing the Knowable from the Unknowable with Language Models
Feb 05, 2024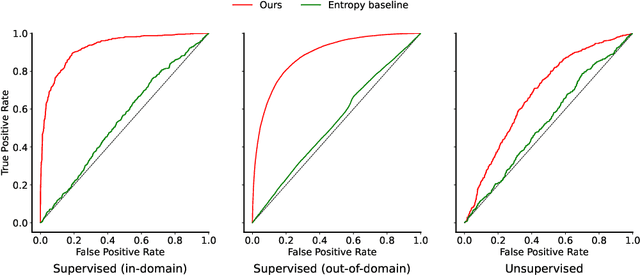
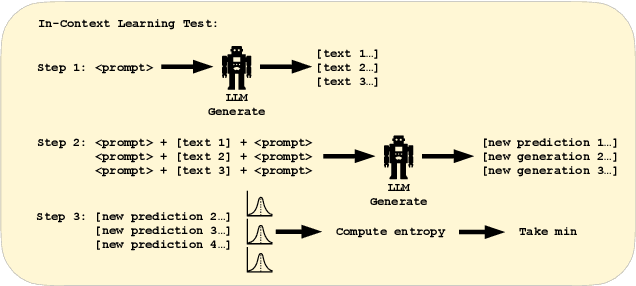

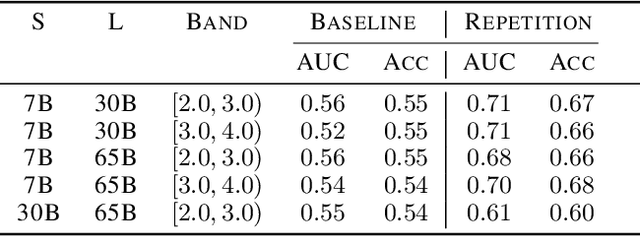
Abstract:We study the feasibility of identifying epistemic uncertainty (reflecting a lack of knowledge), as opposed to aleatoric uncertainty (reflecting entropy in the underlying distribution), in the outputs of large language models (LLMs) over free-form text. In the absence of ground-truth probabilities, we explore a setting where, in order to (approximately) disentangle a given LLM's uncertainty, a significantly larger model stands in as a proxy for the ground truth. We show that small linear probes trained on the embeddings of frozen, pretrained models accurately predict when larger models will be more confident at the token level and that probes trained on one text domain generalize to others. Going further, we propose a fully unsupervised method that achieves non-trivial accuracy on the same task. Taken together, we interpret these results as evidence that LLMs naturally contain internal representations of different types of uncertainty that could potentially be leveraged to devise more informative indicators of model confidence in diverse practical settings.
DeepSpeed4Science Initiative: Enabling Large-Scale Scientific Discovery through Sophisticated AI System Technologies
Oct 11, 2023



Abstract:In the upcoming decade, deep learning may revolutionize the natural sciences, enhancing our capacity to model and predict natural occurrences. This could herald a new era of scientific exploration, bringing significant advancements across sectors from drug development to renewable energy. To answer this call, we present DeepSpeed4Science initiative (deepspeed4science.ai) which aims to build unique capabilities through AI system technology innovations to help domain experts to unlock today's biggest science mysteries. By leveraging DeepSpeed's current technology pillars (training, inference and compression) as base technology enablers, DeepSpeed4Science will create a new set of AI system technologies tailored for accelerating scientific discoveries by addressing their unique complexity beyond the common technical approaches used for accelerating generic large language models (LLMs). In this paper, we showcase the early progress we made with DeepSpeed4Science in addressing two of the critical system challenges in structural biology research.
OpenProteinSet: Training data for structural biology at scale
Aug 10, 2023


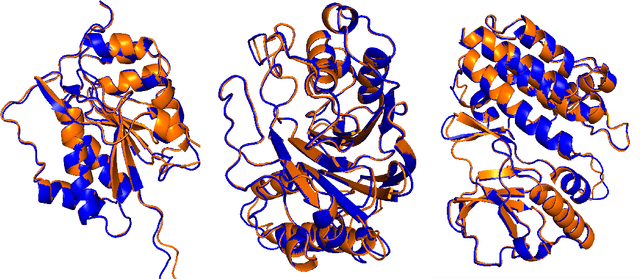
Abstract:Multiple sequence alignments (MSAs) of proteins encode rich biological information and have been workhorses in bioinformatic methods for tasks like protein design and protein structure prediction for decades. Recent breakthroughs like AlphaFold2 that use transformers to attend directly over large quantities of raw MSAs have reaffirmed their importance. Generation of MSAs is highly computationally intensive, however, and no datasets comparable to those used to train AlphaFold2 have been made available to the research community, hindering progress in machine learning for proteins. To remedy this problem, we introduce OpenProteinSet, an open-source corpus of more than 16 million MSAs, associated structural homologs from the Protein Data Bank, and AlphaFold2 protein structure predictions. We have previously demonstrated the utility of OpenProteinSet by successfully retraining AlphaFold2 on it. We expect OpenProteinSet to be broadly useful as training and validation data for 1) diverse tasks focused on protein structure, function, and design and 2) large-scale multimodal machine learning research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge