Benjamin J. Lengerich
Interpretable Predictive Models to Understand Risk Factors for Maternal and Fetal Outcomes
Oct 16, 2023Abstract:Although most pregnancies result in a good outcome, complications are not uncommon and can be associated with serious implications for mothers and babies. Predictive modeling has the potential to improve outcomes through better understanding of risk factors, heightened surveillance for high risk patients, and more timely and appropriate interventions, thereby helping obstetricians deliver better care. We identify and study the most important risk factors for four types of pregnancy complications: (i) severe maternal morbidity, (ii) shoulder dystocia, (iii) preterm preeclampsia, and (iv) antepartum stillbirth. We use an Explainable Boosting Machine (EBM), a high-accuracy glass-box learning method, for prediction and identification of important risk factors. We undertake external validation and perform an extensive robustness analysis of the EBM models. EBMs match the accuracy of other black-box ML methods such as deep neural networks and random forests, and outperform logistic regression, while being more interpretable. EBMs prove to be robust. The interpretability of the EBM models reveals surprising insights into the features contributing to risk (e.g. maternal height is the second most important feature for shoulder dystocia) and may have potential for clinical application in the prediction and prevention of serious complications in pregnancy.
Contextualized Policy Recovery: Modeling and Interpreting Medical Decisions with Adaptive Imitation Learning
Oct 11, 2023Abstract:Interpretable policy learning seeks to estimate intelligible decision policies from observed actions; however, existing models fall short by forcing a tradeoff between accuracy and interpretability. This tradeoff limits data-driven interpretations of human decision-making process. e.g. to audit medical decisions for biases and suboptimal practices, we require models of decision processes which provide concise descriptions of complex behaviors. Fundamentally, existing approaches are burdened by this tradeoff because they represent the underlying decision process as a universal policy, when in fact human decisions are dynamic and can change drastically with contextual information. Thus, we propose Contextualized Policy Recovery (CPR), which re-frames the problem of modeling complex decision processes as a multi-task learning problem in which complex decision policies are comprised of context-specific policies. CPR models each context-specific policy as a linear observation-to-action mapping, and generates new decision models $\textit{on-demand}$ as contexts are updated with new observations. CPR is compatible with fully offline and partially observable decision environments, and can be tailored to incorporate any recurrent black-box model or interpretable decision model. We assess CPR through studies on simulated and real data, achieving state-of-the-art performance on the canonical tasks of predicting antibiotic prescription in intensive care units ($+22\%$ AUROC vs. previous SOTA) and predicting MRI prescription for Alzheimer's patients ($+7.7\%$ AUROC vs. previous SOTA). With this improvement in predictive performance, CPR closes the accuracy gap between interpretable and black-box methods for policy learning, allowing high-resolution exploration and analysis of context-specific decision models.
LLMs Understand Glass-Box Models, Discover Surprises, and Suggest Repairs
Aug 07, 2023Abstract:We show that large language models (LLMs) are remarkably good at working with interpretable models that decompose complex outcomes into univariate graph-represented components. By adopting a hierarchical approach to reasoning, LLMs can provide comprehensive model-level summaries without ever requiring the entire model to fit in context. This approach enables LLMs to apply their extensive background knowledge to automate common tasks in data science such as detecting anomalies that contradict prior knowledge, describing potential reasons for the anomalies, and suggesting repairs that would remove the anomalies. We use multiple examples in healthcare to demonstrate the utility of these new capabilities of LLMs, with particular emphasis on Generalized Additive Models (GAMs). Finally, we present the package $\texttt{TalkToEBM}$ as an open-source LLM-GAM interface.
Using Interpretable Machine Learning to Predict Maternal and Fetal Outcomes
Jul 12, 2022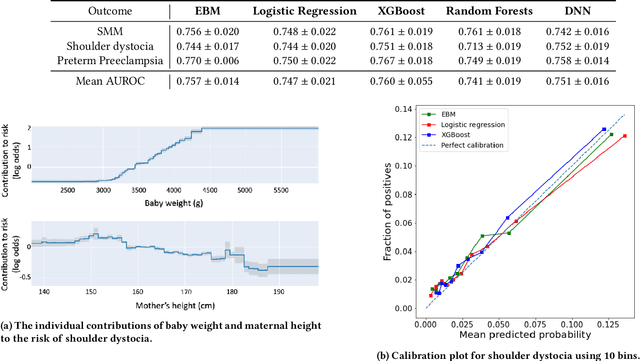

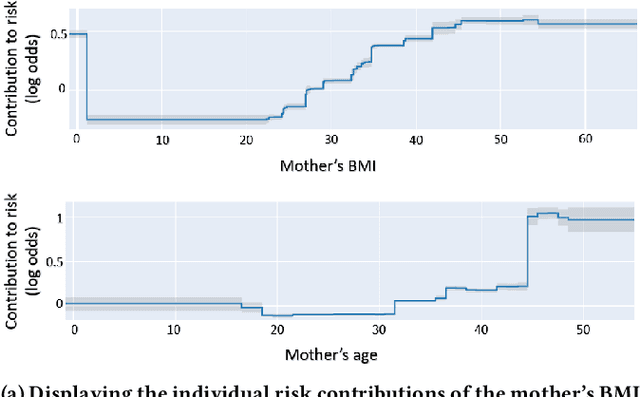
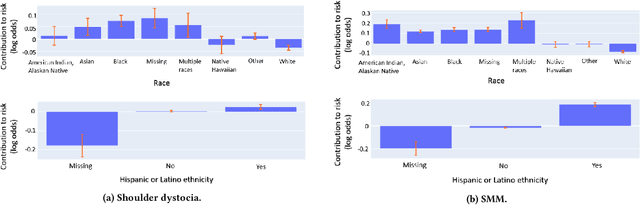
Abstract:Most pregnancies and births result in a good outcome, but complications are not uncommon and when they do occur, they can be associated with serious implications for mothers and babies. Predictive modeling has the potential to improve outcomes through better understanding of risk factors, heightened surveillance, and more timely and appropriate interventions, thereby helping obstetricians deliver better care. For three types of complications we identify and study the most important risk factors using Explainable Boosting Machine (EBM), a glass box model, in order to gain intelligibility: (i) Severe Maternal Morbidity (SMM), (ii) shoulder dystocia, and (iii) preterm preeclampsia. While using the interpretability of EBM's to reveal surprising insights into the features contributing to risk, our experiments show EBMs match the accuracy of other black-box ML methods such as deep neural nets and random forests.
Ten Quick Tips for Deep Learning in Biology
May 29, 2021
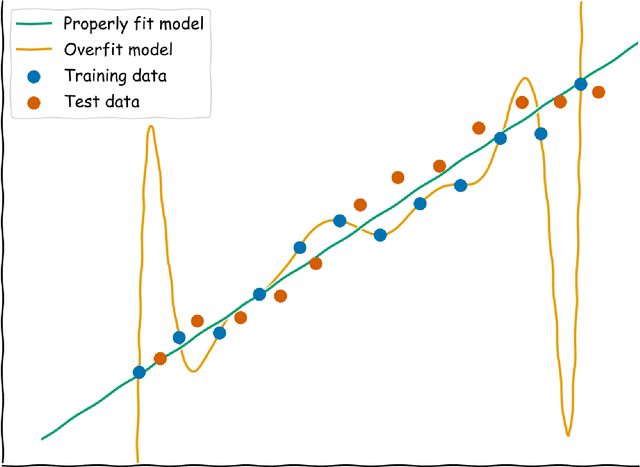
Abstract:Machine learning is a modern approach to problem-solving and task automation. In particular, machine learning is concerned with the development and applications of algorithms that can recognize patterns in data and use them for predictive modeling. Artificial neural networks are a particular class of machine learning algorithms and models that evolved into what is now described as deep learning. Given the computational advances made in the last decade, deep learning can now be applied to massive data sets and in innumerable contexts. Therefore, deep learning has become its own subfield of machine learning. In the context of biological research, it has been increasingly used to derive novel insights from high-dimensional biological data. To make the biological applications of deep learning more accessible to scientists who have some experience with machine learning, we solicited input from a community of researchers with varied biological and deep learning interests. These individuals collaboratively contributed to this manuscript's writing using the GitHub version control platform and the Manubot manuscript generation toolset. The goal was to articulate a practical, accessible, and concise set of guidelines and suggestions to follow when using deep learning. In the course of our discussions, several themes became clear: the importance of understanding and applying machine learning fundamentals as a baseline for utilizing deep learning, the necessity for extensive model comparisons with careful evaluation, and the need for critical thought in interpreting results generated by deep learning, among others.
Hybrid Subspace Learning for High-Dimensional Data
Aug 05, 2018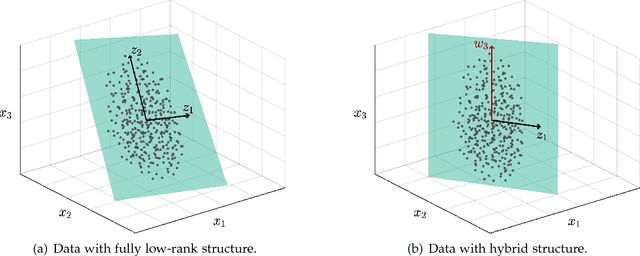
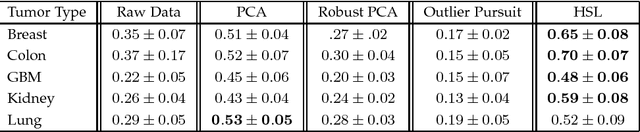
Abstract:The high-dimensional data setting, in which p >> n, is a challenging statistical paradigm that appears in many real-world problems. In this setting, learning a compact, low-dimensional representation of the data can substantially help distinguish signal from noise. One way to achieve this goal is to perform subspace learning to estimate a small set of latent features that capture the majority of the variance in the original data. Most existing subspace learning models, such as PCA, assume that the data can be fully represented by its embedding in one or more latent subspaces. However, in this work, we argue that this assumption is not suitable for many high-dimensional datasets; often only some variables can easily be projected to a low-dimensional space. We propose a hybrid dimensionality reduction technique in which some features are mapped to a low-dimensional subspace while others remain in the original space. Our model leads to more accurate estimation of the latent space and lower reconstruction error. We present a simple optimization procedure for the resulting biconvex problem and show synthetic data results that demonstrate the advantages of our approach over existing methods. Finally, we demonstrate the effectiveness of this method for extracting meaningful features from both gene expression and video background subtraction datasets.
Retrofitting Distributional Embeddings to Knowledge Graphs with Functional Relations
Jun 16, 2018
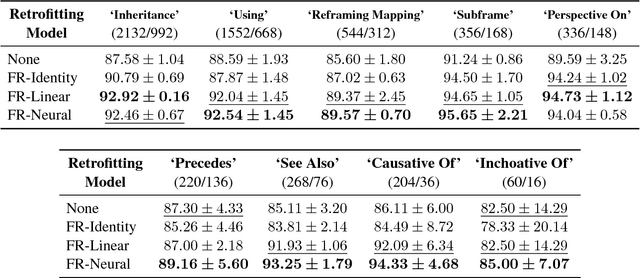
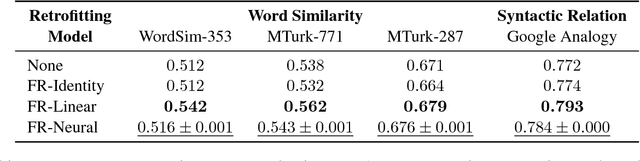
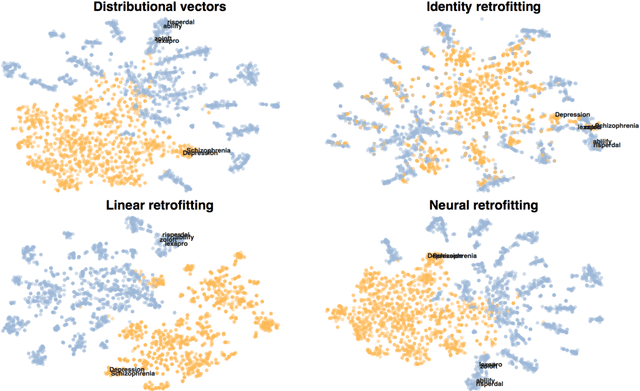
Abstract:Knowledge graphs are a versatile framework to encode richly structured data relationships, but it can be challenging to combine these graphs with unstructured data. Methods for retrofitting pre-trained entity representations to the structure of a knowledge graph typically assume that entities are embedded in a connected space and that relations imply similarity. However, useful knowledge graphs often contain diverse entities and relations (with potentially disjoint underlying corpora) which do not accord with these assumptions. To overcome these limitations, we present Functional Retrofitting, a framework that generalizes current retrofitting methods by explicitly modeling pairwise relations. Our framework can directly incorporate a variety of pairwise penalty functions previously developed for knowledge graph completion. Further, it allows users to encode, learn, and extract information about relation semantics. We present both linear and neural instantiations of the framework. Functional Retrofitting significantly outperforms existing retrofitting methods on complex knowledge graphs and loses no accuracy on simpler graphs (in which relations do imply similarity). Finally, we demonstrate the utility of the framework by predicting new drug--disease treatment pairs in a large, complex health knowledge graph.
Towards Visual Explanations for Convolutional Neural Networks via Input Resampling
Aug 16, 2017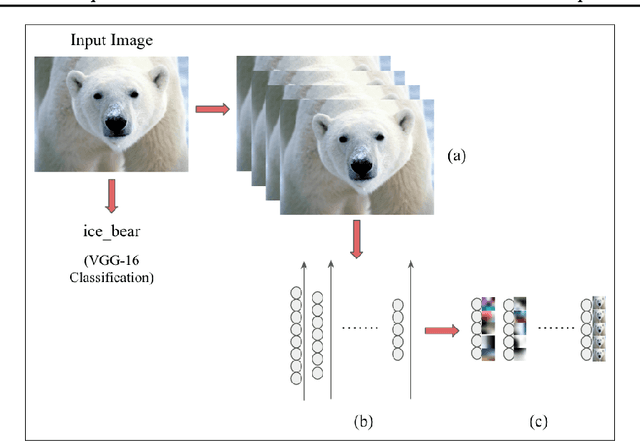
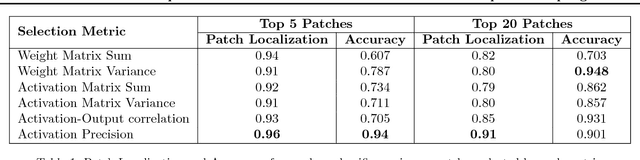
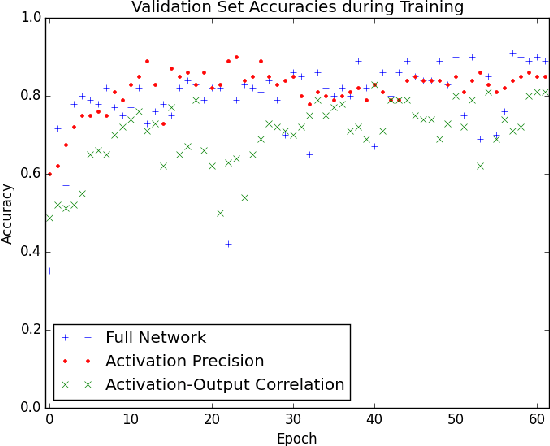
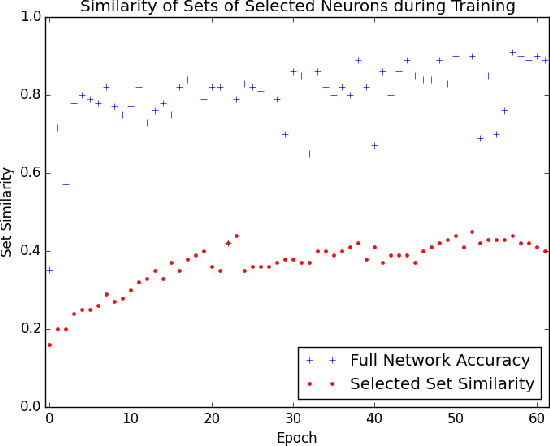
Abstract:The predictive power of neural networks often costs model interpretability. Several techniques have been developed for explaining model outputs in terms of input features; however, it is difficult to translate such interpretations into actionable insight. Here, we propose a framework to analyze predictions in terms of the model's internal features by inspecting information flow through the network. Given a trained network and a test image, we select neurons by two metrics, both measured over a set of images created by perturbations to the input image: (1) magnitude of the correlation between the neuron activation and the network output and (2) precision of the neuron activation. We show that the former metric selects neurons that exert large influence over the network output while the latter metric selects neurons that activate on generalizable features. By comparing the sets of neurons selected by these two metrics, our framework suggests a way to investigate the internal attention mechanisms of convolutional neural networks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge