Yin Aphinyanaphongs
LLMs Understand Glass-Box Models, Discover Surprises, and Suggest Repairs
Aug 07, 2023Abstract:We show that large language models (LLMs) are remarkably good at working with interpretable models that decompose complex outcomes into univariate graph-represented components. By adopting a hierarchical approach to reasoning, LLMs can provide comprehensive model-level summaries without ever requiring the entire model to fit in context. This approach enables LLMs to apply their extensive background knowledge to automate common tasks in data science such as detecting anomalies that contradict prior knowledge, describing potential reasons for the anomalies, and suggesting repairs that would remove the anomalies. We use multiple examples in healthcare to demonstrate the utility of these new capabilities of LLMs, with particular emphasis on Generalized Additive Models (GAMs). Finally, we present the package $\texttt{TalkToEBM}$ as an open-source LLM-GAM interface.
Diagnosis Uncertain Models For Medical Risk Prediction
Jun 29, 2023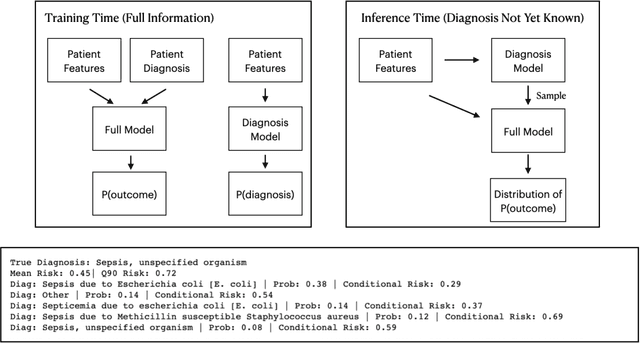
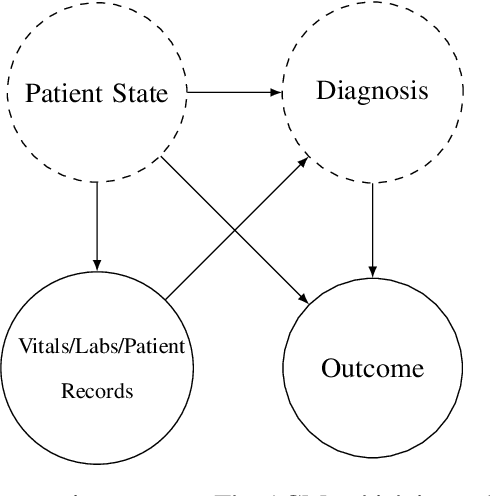
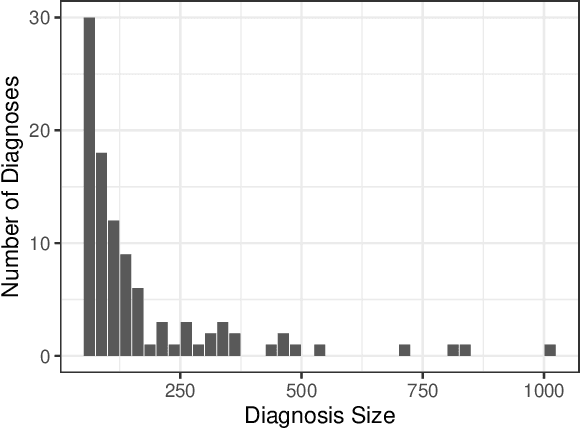
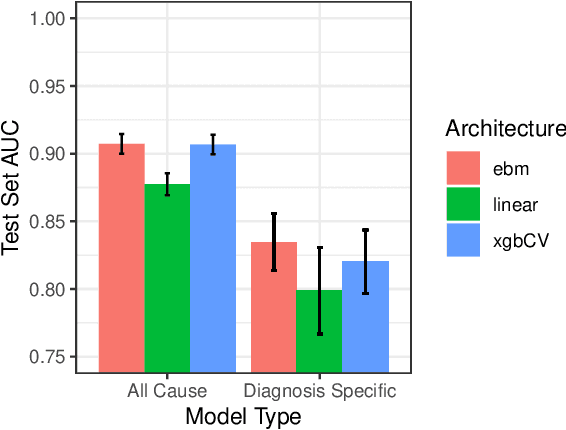
Abstract:We consider a patient risk models which has access to patient features such as vital signs, lab values, and prior history but does not have access to a patient's diagnosis. For example, this occurs in a model deployed at intake time for triage purposes. We show that such `all-cause' risk models have good generalization across diagnoses but have a predictable failure mode. When the same lab/vital/history profiles can result from diagnoses with different risk profiles (e.g. E.coli vs. MRSA) the risk estimate is a probability weighted average of these two profiles. This leads to an under-estimation of risk for rare but highly risky diagnoses. We propose a fix for this problem by explicitly modeling the uncertainty in risk prediction coming from uncertainty in patient diagnoses. This gives practitioners an interpretable way to understand patient risk beyond a single risk number.
Estimating Discontinuous Time-Varying Risk Factors and Treatment Benefits for COVID-19 with Interpretable ML
Nov 15, 2022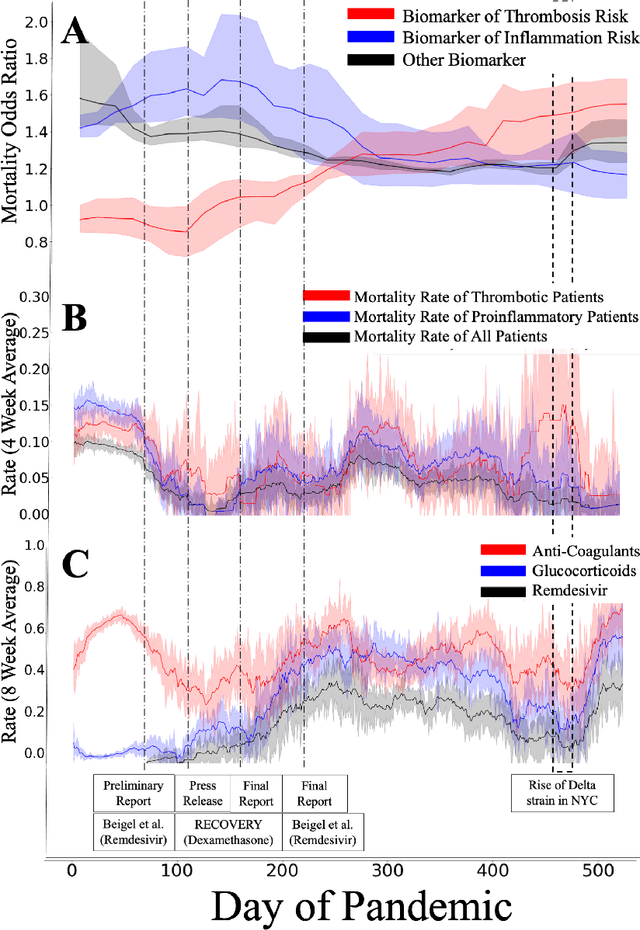
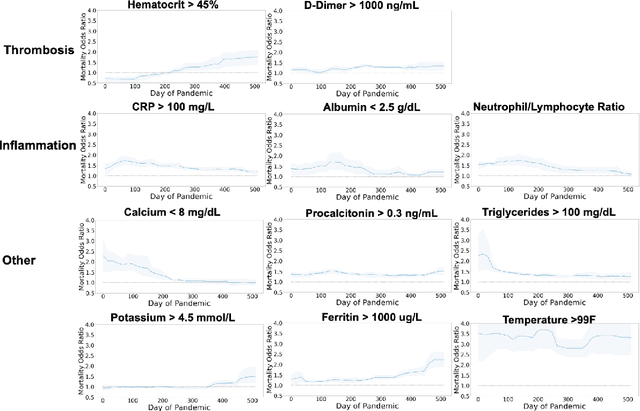
Abstract:Treatment protocols, disease understanding, and viral characteristics changed over the course of the COVID-19 pandemic; as a result, the risks associated with patient comorbidities and biomarkers also changed. We add to the conversation regarding inflammation, hemostasis and vascular function in COVID-19 by performing a time-varying observational analysis of over 4000 patients hospitalized for COVID-19 in a New York City hospital system from March 2020 to August 2021. To perform this analysis, we apply tree-based generalized additive models with temporal interactions which recover discontinuous risk changes caused by discrete protocols changes. We find that the biomarkers of thrombosis increasingly predicted mortality from March 2020 to August 2021, while the association between biomarkers of inflammation and thrombosis weakened. Beyond COVID-19, this presents a straightforward methodology to estimate unknown and discontinuous time-varying effects.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge