Diagnosis Uncertain Models For Medical Risk Prediction
Paper and Code
Jun 29, 2023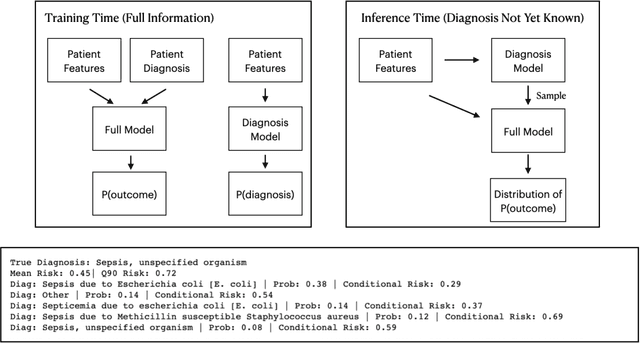
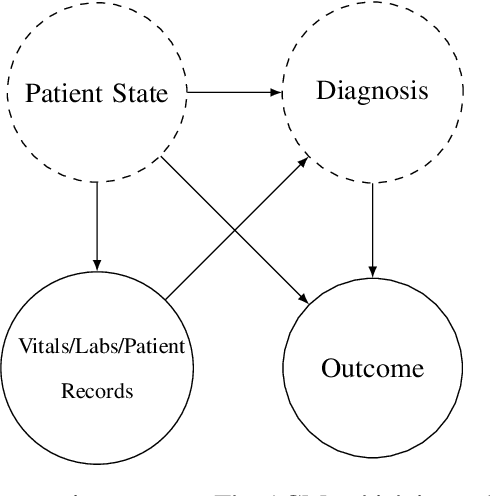
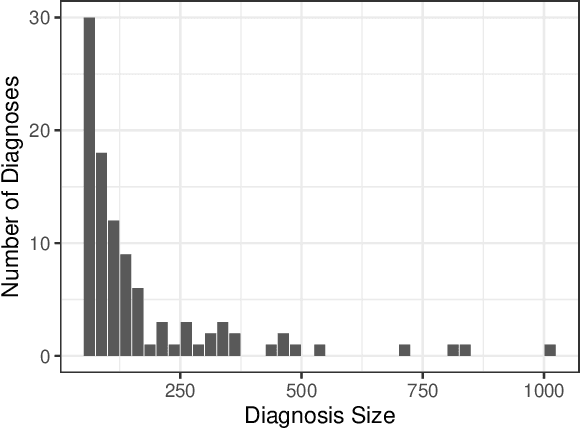
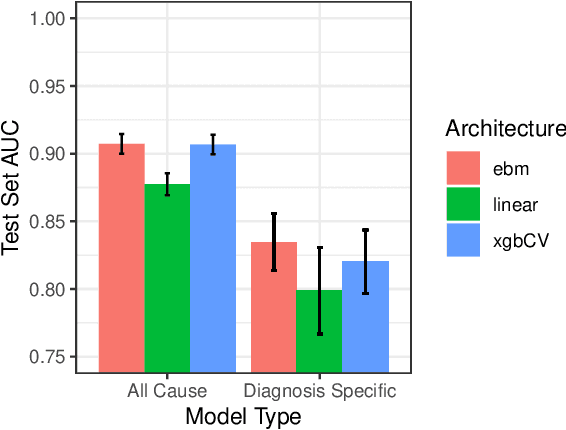
We consider a patient risk models which has access to patient features such as vital signs, lab values, and prior history but does not have access to a patient's diagnosis. For example, this occurs in a model deployed at intake time for triage purposes. We show that such `all-cause' risk models have good generalization across diagnoses but have a predictable failure mode. When the same lab/vital/history profiles can result from diagnoses with different risk profiles (e.g. E.coli vs. MRSA) the risk estimate is a probability weighted average of these two profiles. This leads to an under-estimation of risk for rare but highly risky diagnoses. We propose a fix for this problem by explicitly modeling the uncertainty in risk prediction coming from uncertainty in patient diagnoses. This gives practitioners an interpretable way to understand patient risk beyond a single risk number.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge