Asheesh K. Singh
Iowa State University
Soybean Maturity Prediction using 2D Contour Plots from Drone based Time Series Imagery
Dec 12, 2024Abstract:Plant breeding programs require assessments of days to maturity for accurate selection and placement of entries in appropriate tests. In the early stages of the breeding pipeline, soybean breeding programs assign relative maturity ratings to experimental varieties that indicate their suitable maturity zones. Traditionally, the estimation of maturity value for breeding varieties has involved breeders manually inspecting fields and assessing maturity value visually. This approach relies heavily on rater judgment, making it subjective and time-consuming. This study aimed to develop a machine-learning model for evaluating soybean maturity using UAV-based time-series imagery. Images were captured at three-day intervals, beginning as the earliest varieties started maturing and continuing until the last varieties fully matured. The data collected for this experiment consisted of 22,043 plots collected across three years (2021 to 2023) and represent relative maturity groups 1.6 - 3.9. We utilized contour plot images extracted from the time-series UAV RGB imagery as input for a neural network model. This contour plot approach encoded the temporal and spatial variation within each plot into a single image. A deep learning model was trained to utilize this contour plot to predict maturity ratings. This model significantly improves accuracy and robustness, achieving up to 85% accuracy. We also evaluate the model's accuracy as we reduce the number of time points, quantifying the trade-off between temporal resolution and maturity prediction. The predictive model offers a scalable, objective, and efficient means of assessing crop maturity, enabling phenomics and ML approaches to reduce the reliance on manual inspection and subjective assessment. This approach enables the automatic prediction of relative maturity ratings in a breeding program, saving time and resources.
AgEval: A Benchmark for Zero-Shot and Few-Shot Plant Stress Phenotyping with Multimodal LLMs
Jul 29, 2024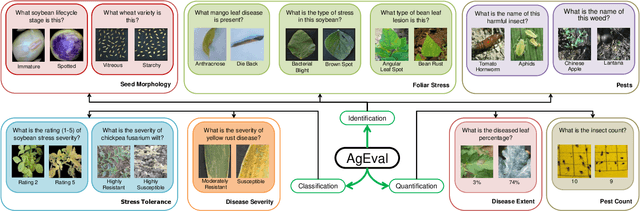
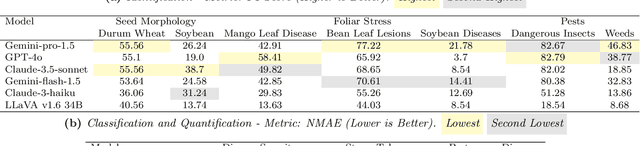
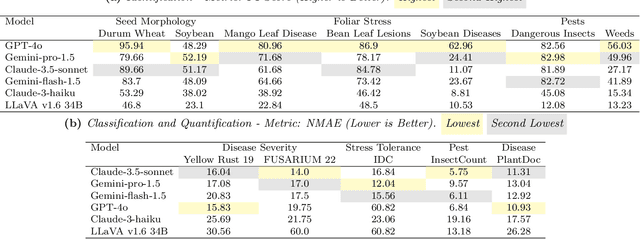
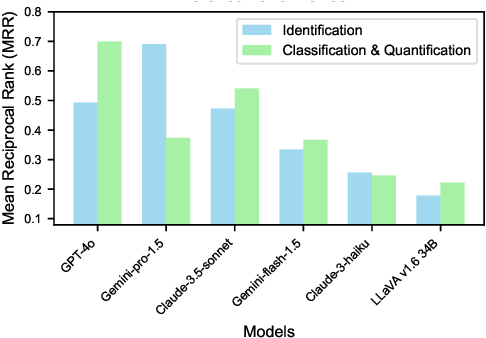
Abstract:Plant stress phenotyping traditionally relies on expert assessments and specialized models, limiting scalability in agriculture. Recent advances in multimodal large language models (LLMs) offer potential solutions to this challenge. We present AgEval, a benchmark comprising 12 diverse plant stress phenotyping tasks, to evaluate these models' capabilities. Our study assesses zero-shot and few-shot in-context learning performance of state-of-the-art models, including Claude, GPT, Gemini, and LLaVA. Results show significant performance improvements with few-shot learning, with F1 scores increasing from 46.24% to 73.37% in 8-shot identification for the best-performing model. Few-shot examples from other classes in the dataset have negligible or negative impacts, although having the exact category example helps to increase performance by 15.38%. We also quantify the consistency of model performance across different classes within each task, finding that the coefficient of variance (CV) ranges from 26.02% to 58.03% across models, implying that subject matter expertise is needed - of 'difficult' classes - to achieve reliability in performance. AgEval establishes baseline metrics for multimodal LLMs in agricultural applications, offering insights into their promise for enhancing plant stress phenotyping at scale. Benchmark and code can be accessed at: https://anonymous.4open.science/r/AgEval/
Class-specific Data Augmentation for Plant Stress Classification
Jun 18, 2024
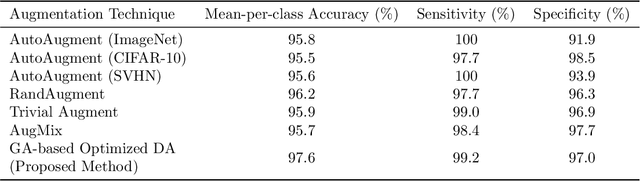
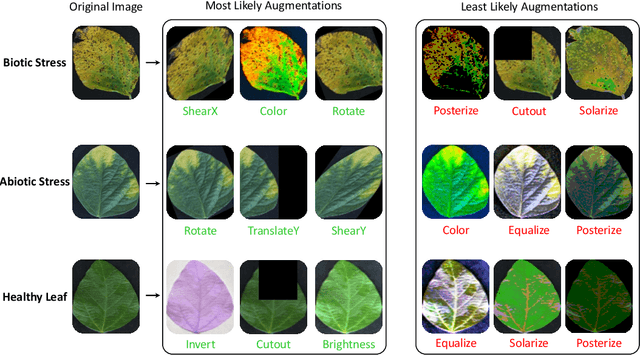
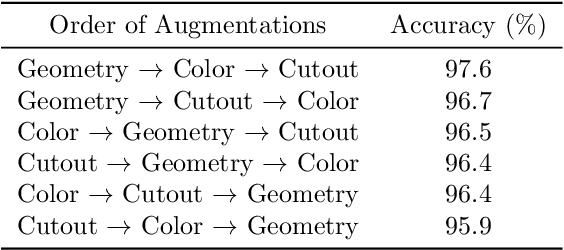
Abstract:Data augmentation is a powerful tool for improving deep learning-based image classifiers for plant stress identification and classification. However, selecting an effective set of augmentations from a large pool of candidates remains a key challenge, particularly in imbalanced and confounding datasets. We propose an approach for automated class-specific data augmentation using a genetic algorithm. We demonstrate the utility of our approach on soybean [Glycine max (L.) Merr] stress classification where symptoms are observed on leaves; a particularly challenging problem due to confounding classes in the dataset. Our approach yields substantial performance, achieving a mean-per-class accuracy of 97.61% and an overall accuracy of 98% on the soybean leaf stress dataset. Our method significantly improves the accuracy of the most challenging classes, with notable enhancements from 83.01% to 88.89% and from 85.71% to 94.05%, respectively. A key observation we make in this study is that high-performing augmentation strategies can be identified in a computationally efficient manner. We fine-tune only the linear layer of the baseline model with different augmentations, thereby reducing the computational burden associated with training classifiers from scratch for each augmentation policy while achieving exceptional performance. This research represents an advancement in automated data augmentation strategies for plant stress classification, particularly in the context of confounding datasets. Our findings contribute to the growing body of research in tailored augmentation techniques and their potential impact on disease management strategies, crop yields, and global food security. The proposed approach holds the potential to enhance the accuracy and efficiency of deep learning-based tools for managing plant stresses in agriculture.
Multi-Sensor and Multi-temporal High-Throughput Phenotyping for Monitoring and Early Detection of Water-Limiting Stress in Soybean
Feb 28, 2024Abstract:Soybean production is susceptible to biotic and abiotic stresses, exacerbated by extreme weather events. Water limiting stress, i.e. drought, emerges as a significant risk for soybean production, underscoring the need for advancements in stress monitoring for crop breeding and production. This project combines multi-modal information to identify the most effective and efficient automated methods to investigate drought response. We investigated a set of diverse soybean accessions using multiple sensors in a time series high-throughput phenotyping manner to: (1) develop a pipeline for rapid classification of soybean drought stress symptoms, and (2) investigate methods for early detection of drought stress. We utilized high-throughput time-series phenotyping using UAVs and sensors in conjunction with machine learning (ML) analytics, which offered a swift and efficient means of phenotyping. The red-edge and green bands were most effective to classify canopy wilting stress. The Red-Edge Chlorophyll Vegetation Index (RECI) successfully differentiated susceptible and tolerant soybean accessions prior to visual symptom development. We report pre-visual detection of soybean wilting using a combination of different vegetation indices. These results can contribute to early stress detection methodologies and rapid classification of drought responses in screening nurseries for breeding and production applications.
Deep Multi-view Image Fusion for Soybean Yield Estimation in Breeding Applications Deep Multi-view Image Fusion for Soybean Yield Estimation in Breeding Applications
Nov 13, 2020
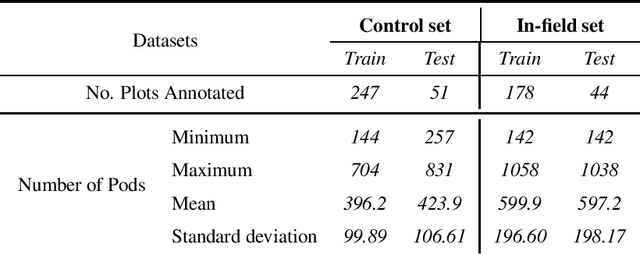
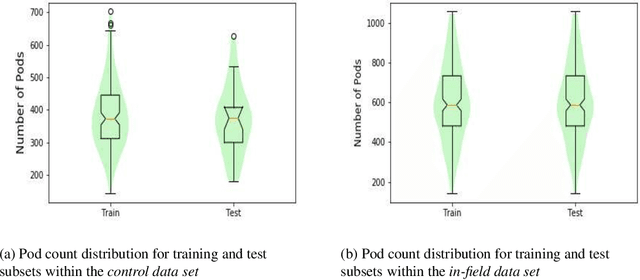
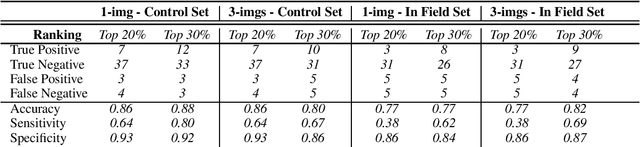
Abstract:Reliable seed yield estimation is an indispensable step in plant breeding programs geared towards cultivar development in major row crops. The objective of this study is to develop a machine learning (ML) approach adept at soybean [\textit{Glycine max} L. (Merr.)] pod counting to enable genotype seed yield rank prediction from in-field video data collected by a ground robot. To meet this goal, we developed a multi-view image-based yield estimation framework utilizing deep learning architectures. Plant images captured from different angles were fused to estimate the yield and subsequently to rank soybean genotypes for application in breeding decisions. We used data from controlled imaging environment in field, as well as from plant breeding test plots in field to demonstrate the efficacy of our framework via comparing performance with manual pod counting and yield estimation. Our results demonstrate the promise of ML models in making breeding decisions with significant reduction of time and human effort, and opening new breeding methods avenues to develop cultivars.
Usefulness of interpretability methods to explain deep learning based plant stress phenotyping
Jul 11, 2020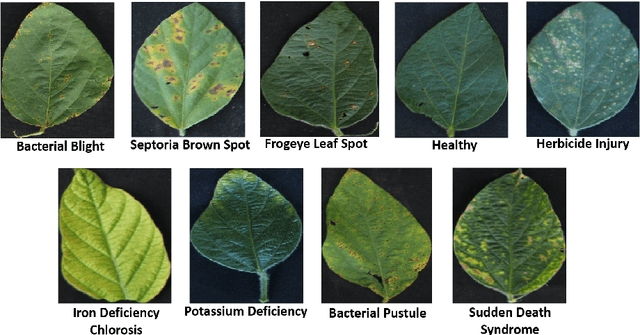



Abstract:Deep learning techniques have been successfully deployed for automating plant stress identification and quantification. In recent years, there is a growing push towards training models that are interpretable -i.e. that justify their classification decisions by visually highlighting image features that were crucial for classification decisions. The expectation is that trained network models utilize image features that mimic visual cues used by plant pathologists. In this work, we compare some of the most popular interpretability methods: Saliency Maps, SmoothGrad, Guided Backpropogation, Deep Taylor Decomposition, Integrated Gradients, Layer-wise Relevance Propagation and Gradient times Input, for interpreting the deep learning model. We train a DenseNet-121 network for the classification of eight different soybean stresses (biotic and abiotic). Using a dataset consisting of 16,573 RGB images of healthy and stressed soybean leaflets captured under controlled conditions, we obtained an overall classification accuracy of 95.05 \%. For a diverse subset of the test data, we compared the important features with those identified by a human expert. We observed that most interpretability methods identify the infected regions of the leaf as important features for some -- but not all -- of the correctly classified images. For some images, the output of the interpretability methods indicated that spurious feature correlations may have been used to correctly classify them. Although the output explanation maps of these interpretability methods may be different from each other for a given image, we advocate the use of these interpretability methods as `hypothesis generation' mechanisms that can drive scientific insight.
How useful is Active Learning for Image-based Plant Phenotyping?
Jul 01, 2020



Abstract:Deep learning models have been successfully deployed for a diverse array of image-based plant phenotyping applications including disease detection and classification. However, successful deployment of supervised deep learning models requires large amount of labeled data, which is a significant challenge in plant science (and most biological) domains due to the inherent complexity. Specifically, data annotation is costly, laborious, time consuming and needs domain expertise for phenotyping tasks, especially for diseases. To overcome this challenge, active learning algorithms have been proposed that reduce the amount of labeling needed by deep learning models to achieve good predictive performance. Active learning methods adaptively select samples to annotate using an acquisition function to achieve maximum (classification) performance under a fixed labeling budget. We report the performance of four different active learning methods, (1) Deep Bayesian Active Learning (DBAL), (2) Entropy, (3) Least Confidence, and (4) Coreset, with conventional random sampling-based annotation for two different image-based classification datasets. The first image dataset consists of soybean [Glycine max L. (Merr.)] leaves belonging to eight different soybean stresses and a healthy class, and the second consists of nine different weed species from the field. For a fixed labeling budget, we observed that the classification performance of deep learning models with active learning-based acquisition strategies is better than random sampling-based acquisition for both datasets. The integration of active learning strategies for data annotation can help mitigate labelling challenges in the plant sciences applications particularly where deep domain knowledge is required.
Crop Yield Prediction Integrating Genotype and Weather Variables Using Deep Learning
Jun 24, 2020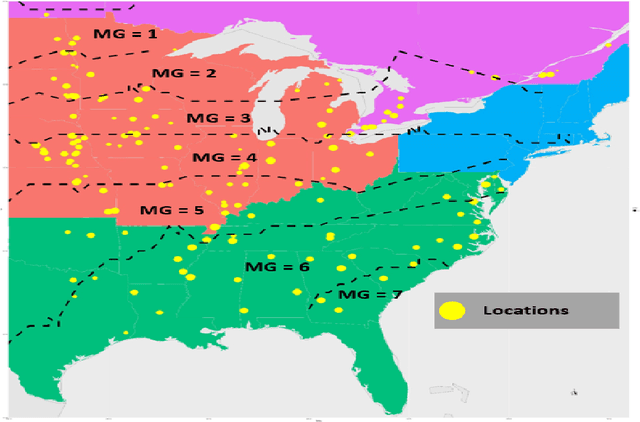
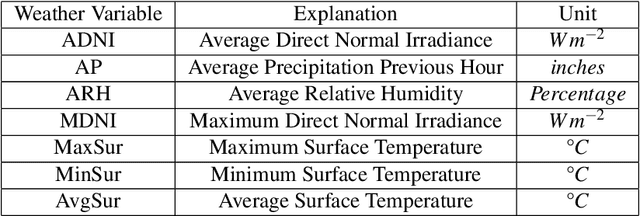
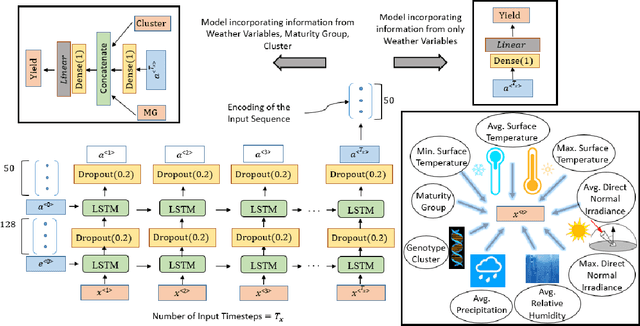

Abstract:Accurate prediction of crop yield supported by scientific and domain-relevant insights, can help improve agricultural breeding, provide monitoring across diverse climatic conditions and thereby protect against climatic challenges to crop production including erratic rainfall and temperature variations. We used historical performance records from Uniform Soybean Tests (UST) in North America spanning 13 years of data to build a Long Short Term Memory - Recurrent Neural Network based model to dissect and predict genotype response in multiple-environments by leveraging pedigree relatedness measures along with weekly weather parameters. Additionally, for providing explainability of the important time-windows in the growing season, we developed a model based on temporal attention mechanism. The combination of these two models outperformed random forest (RF), LASSO regression and the data-driven USDA model for yield prediction. We deployed this deep learning framework as a 'hypotheses generation tool' to unravel GxExM relationships. Attention-based time series models provide a significant advancement in interpretability of yield prediction models. The insights provided by explainable models are applicable in understanding how plant breeding programs can adapt their approaches for global climate change, for example identification of superior varieties for commercial release, intelligent sampling of testing environments in variety development, and integrating weather parameters for a targeted breeding approach. Using DL models as hypothesis generation tools will enable development of varieties with plasticity response in variable climatic conditions. We envision broad applicability of this approach (via conducting sensitivity analysis and "what-if" scenarios) for soybean and other crop species under different climatic conditions.
Explaining hyperspectral imaging based plant disease identification: 3D CNN and saliency maps
Apr 24, 2018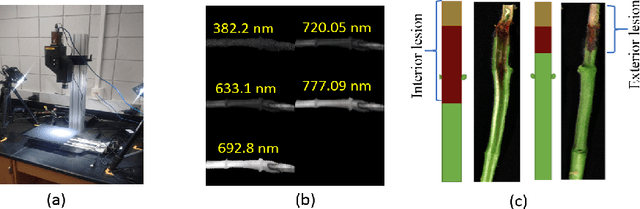
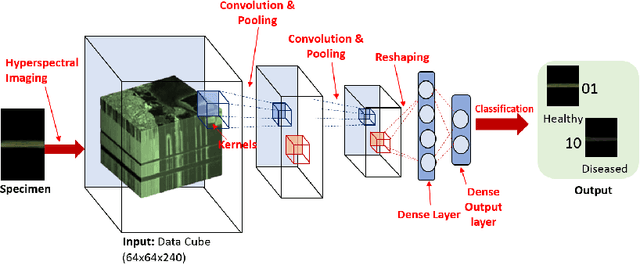


Abstract:Our overarching goal is to develop an accurate and explainable model for plant disease identification using hyperspectral data. Charcoal rot is a soil borne fungal disease that affects the yield of soybean crops worldwide. Hyperspectral images were captured at 240 different wavelengths in the range of 383 - 1032 nm. We developed a 3D Convolutional Neural Network model for soybean charcoal rot disease identification. Our model has classification accuracy of 95.73\% and an infected class F1 score of 0.87. We infer the trained model using saliency map and visualize the most sensitive pixel locations that enable classification. The sensitivity of individual wavelengths for classification was also determined using the saliency map visualization. We identify the most sensitive wavelength as 733 nm using the saliency map visualization. Since the most sensitive wavelength is in the Near Infrared Region(700 - 1000 nm) of the electromagnetic spectrum, which is also the commonly used spectrum region for determining the vegetation health of the plant, we were more confident in the predictions using our model.
Interpretable Deep Learning applied to Plant Stress Phenotyping
Oct 28, 2017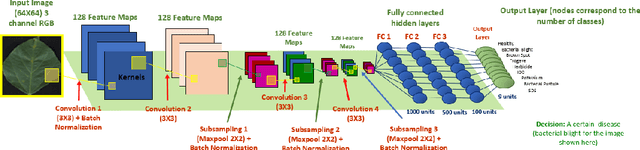
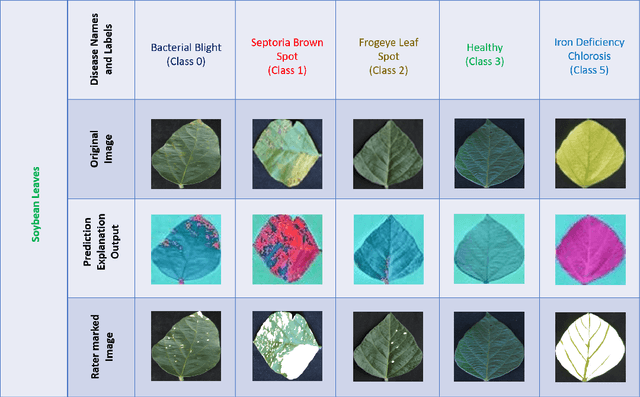
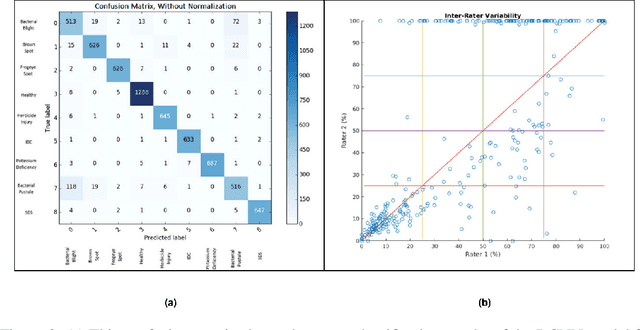
Abstract:Availability of an explainable deep learning model that can be applied to practical real world scenarios and in turn, can consistently, rapidly and accurately identify specific and minute traits in applicable fields of biological sciences, is scarce. Here we consider one such real world example viz., accurate identification, classification and quantification of biotic and abiotic stresses in crop research and production. Up until now, this has been predominantly done manually by visual inspection and require specialized training. However, such techniques are hindered by subjectivity resulting from inter- and intra-rater cognitive variability. Here, we demonstrate the ability of a machine learning framework to identify and classify a diverse set of foliar stresses in the soybean plant with remarkable accuracy. We also present an explanation mechanism using gradient-weighted class activation mapping that isolates the visual symptoms used by the model to make predictions. This unsupervised identification of unique visual symptoms for each stress provides a quantitative measure of stress severity, allowing for identification, classification and quantification in one framework. The learnt model appears to be agnostic to species and make good predictions for other (non-soybean) species, demonstrating an ability of transfer learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge